Back to Journals » Cancer Management and Research » Volume 9
Is the prognostic significance of O6-methylguanine-DNA methyltransferase promoter methylation equally important in glioblastomas of patients from different continents? A systematic review with meta-analysis
Received 26 April 2017
Accepted for publication 14 July 2017
Published 20 September 2017 Volume 2017:9 Pages 411—425
DOI https://doi.org/10.2147/CMAR.S140447
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Alexandra R. Fernandes
Wei Meng,1,* Yangyang Jiang,2,* Jie Ma1
1Department of Pediatric Neurosurgery, Xin Hua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shanghai, 2Department of Neurosurgery, Shanghai Deji Hospital, Shanghai Neuromedical Center Affiliated to Qingdao University, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Background: O6-methylguanine-DNA methyltransferase (MGMT) is an independent predictor of therapeutic response and potential prognosis in patients with glioblastoma multiforme (GBM). However, its significance of clinical prognosis in different continents still needs to be explored.
Patients and methods: To explore the effects of MGMT promoter methylation on both progression-free survival (PFS) and overall survival (OS) among GBM patients from different continents, a systematic review of published studies was conducted.
Results: A total of 5103 patients from 53 studies were involved in the systematic review and the total percentage of MGMT promoter methylation was 45.53%. Of these studies, 16 studies performed univariate analyses and 17 performed multivariate analyses of MGMT promoter methylation on PFS. The pooled hazard ratio (HR) estimated for PFS was 0.55 (95% CI 0.50, 0.60) by univariate analysis and 0.43 (95% CI 0.38, 0.48) by multivariate analysis. The effect of MGMT promoter methylation on OS was explored in 30 studies by univariate analysis and in 30 studies by multivariate analysis. The combined HR was 0.48 (95% CI 0.44, 0.52) and 0.42 (95% CI 0.38, 0.45), respectively.
Conclusion: In each subgroup divided by areas, the prognostic significance still remained highly significant. The proportion of methylation in each group was in inverse proportion to the corresponding HR in the univariate and multivariate analyses of PFS. However, from the perspective of OS, compared with data from Europe and the US, higher methylation rates in Asia did not bring better returns.
Keywords: O6-methylguanine-DNA methyltransferase, methylation, glioblastoma, prognosis, meta-analysis
Introduction
Glioblastoma multiforme (GBM, WHO grade 4) is the most common primary brain tumor in adults with an annual incidence of 3–4/100,000 and is associated with poor prognosis.1 Although some clinical trials have demonstrated that the standard treatment improves overall survival (OS) and progression-free survival (PFS), only less than one-third of GBM patients seem to benefit from these therapies, mainly because of GBM resistance to alkylating drugs.
Transcriptionally active O6-methylguanine-DNA methyltransferase (MGMT) gene encodes a ubiquitously expressed suicide DNA repair enzyme that counteracts the normally lethal effects of alkylating agents by removing the alkyl adducts, preventing the formation of cross-links and thereby causing resistance to alkylating drugs.3 The loss of MGMT protein expression caused by methylation of the MGMT promoter reduces the DNA repair activity of glioma cells, preventing their resistance to alkylating agents.2,4–6 It is believed that patients with GBM who have a methylated MGMT promoter are more sensitive to the killing effects of alkylating drugs, because tumor cells with low MGMT expression were unable to repair such DNA lesions and, thus, were prone to apoptosis, whereas those that do not have a methylated MGMT promoter do not have this benefit.68,69
Various studies have shown that the MGMT promoter methylation status is an independent prognostic factor to GBM and the assessment of MGMT promoter methylation is currently considered as mandatory for patient selection in clinical trials.7–10,68 However, many differences in high risk factors and postoperative chemoradiation stay in guidelines for the treatment of glioblastoma, among countries, indicating different attitudes to MGMT promoter methylation status. Is the prognostic significance of MGMT promoter methylation independent equally among glioblastomas from different areas? Further explorations are needed in the prognostic value of MGMT promoter methylation on GBM including therapeutic intervention.11,12, 20, 21
From the perspective of geography, we conducted this meta-analysis to test the independence of prognostic value of MGMT promoter methylation in both PFS and OS among patients with GBM.
Patients and methods
Publication selection
Ethical approval and patient consent are not required as this is a systematic review and meta-analysis of previously published studies. This study was performed in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement.13
Two reviewers (Yangyang Jiang and Wei Meng) participated in the citations search, study selection and data extraction, independently. Divergences between reviewers were resolved through consulting with Professor Jie Ma.
Electronic databases, including PubMed, EMBASE, Web of Science, China Biomedical Literature Database (CBM), Chinese National Knowledge Infrastructure (CNKI), China Wan Fang database and the Cochrane library, were searched for relevant clinical trials published on the association between MGMT promoter methylation and GBM between January 2000 and June 2017.
The search combined key words: (“O6-methylguanine-DNA methyltransferase methylation” OR MGMT methylation”) AND (“glioblastoma” OR “GBM”) AND (“survival analysis” OR “meta analysis”) AND (“MSP” OR “PSQ”) AND (“survival analysis” OR “meta analysis”) AND (“methylation-specific polymerase chain reaction and pyrosequencing”).
The meta-analysis gathered complete databases from published cohort studies dealing with the prognostic value of MGMT promoter methylation in patients with GBM no matter which therapy was given.
The language in which the papers were written was restricted to English and Chinese. Abstracts were excluded because of insufficient data for meta-analysis. In order to identify the relevant publications, the references cited in the research papers were also scanned. To avoid duplication of data, we carefully noted the author names and the different research centers involved. We evaluated the eligible studies if all the following conditions were met: 1) MGMT promoter methylation status was measured by using identified method such as methylation-specific polymerase chain reaction (MSP) and pyrosequencing (PSQ); (2) inclusion of sufficient data or survival curves to calculate hazard ratio (HR) and 95% CI; and 3) full or special parts of papers investigated the relationship between MGMT promoter methylation and PFS or OS.
Data extraction
Two authors (Yangyang Jiang and Wei Meng) independently reviewed and extracted the data needed. Disagreements were resolved through discussion with each other.
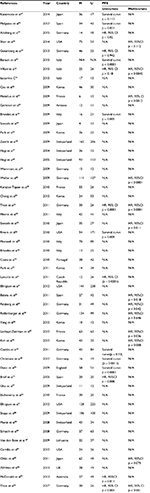
We used a predesigned data extraction sheet to obtain the following information: first author, year of publication, region, HR form and sample size and style of postoperative chemoradiation, if given. The formula recommended by Spruance et al was adopted to calculate the corresponding HR of the missing data.14 Kaplan–Meier curve was read by using Engauge Digitizer version 4.1 (available at: http://sourceforge.net/) except if the paper has supplied HR directly.15 (All the data are shown in Table 1.)
Statistical analysis
In some studies, HR and 95% CI were directly obtained from published literature by using univariate or multivariate survival analysis. For studies in which the HR corresponding to the 95% CI was not given directly, published data and figures from original papers were used to calculate the HR according to the methods described by using Engauge Digitizer version 4.1.
The pooled HR corresponding to the 95% CI was used to assess the prognostic value of MGMT promoter methylation in patients with GBM. The statistical heterogeneity among studies was assessed with the Q-test and I2 statistics.16
A random-effects model was used primarily regardless of heterogeneity. Level of heterogeneity (level of variance) across studies was evaluated using I2 statistic. I2 of 40, 70 and 100% was used to represent low, moderate and high variance, respectively.17 If obvious differences for clinical characteristics and methodology were not identified and I2 ≤ 40%, a fixed-effects model was adopted. A random-effects model will be used if clinical characteristics and methodology were not identified to be great difference and I2 ≤ 40%; in contrast, if the clinical characteristic and/or methodology across studies regardless of I2 statistic was considered to be obviously different, qualitative analysis was adopted.18
The objective impact of MGMT promoter methylation on PFS and OS was considered to be statistically significant if the 95% CI for the HR did not overlap 0. Publication bias was evaluated with funnel plot and Begg’s rank correlation method.19 The statistical analyses were performed by STATA/MP 13.0 software.
Results
Characteristics of studies
A total of 204 relevant citations were identified at the initial search stage; 151 articles concerned topics not relevant to this study, and finally 53 studies were included in the meta-analysis.
All the included studies were in English. The individual characteristics of the eligible studies are reported in Table 1. The total number of patients was 5103, and the total frequency of MGMT promoter methylation was 45.53%. Of the 53 publications eligible for systematic review, 31 studies reported the HR with corresponding to 95% CI directly, the other 22 studies reported the HR in the style of survival curve availability.
Meta-analysis
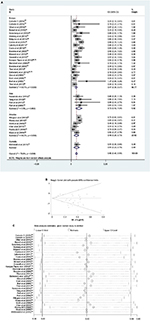
Sixteen studies (one in Asia, one in North America, one in Australia and 13 in Europe) reported the effect of MGMT promoter methylation on PFS using univariate analysis.12,23–25,31,44,46–49,56,59,68,73,79,93 As shown in Figure 1, the HR of the Asian group is 0.47, the HR of the American group is 0.88, the HR of the Australian group is 0.51 and the HR of the European group is 0.49; MGMT promoter methylation was significantly correlated with better PFS according to univariate analysis, with a combined HR of 0.55 (95% CI 0.50, 0.60).The random-effects model (the DerSimonian and Laird method) was used because significant heterogeneity was detected among these studies (p = 0.000, I2 = 88.3%).61
The effect of MGMT promoter methylation on PFS adjusted for other variables was evaluated in 17 studies (five in Asia, 11 in Europe and one in America)26–30,32,34,35,41,45,68,85,87,91,93–95 As shown in Figure 2, the HR of the Asian group is 0.49, the HR of the European group is 0.44 and the HR of the American group is 0.37; MGMT promoter methylation was significantly correlated with better PFS according to multivariate analysis, with a combined HR of 0.45 (95% CI 0.35, 0.54). The random-effects model (the DerSimonian and Laird method) was used because significant heterogeneity was detected among these studies (p = 0.000, I2 = 62.8%).61
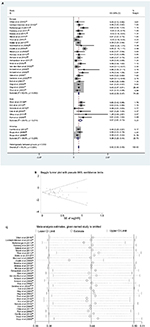
The effect of MGMT promoter methylation on OS unadjusted for using univariate analysis was evaluated in 32 studies (four in Asia, six in North America, one in Australia and 21 in Europe).12,22–25,29,31,34,36,39,43,44,46,47,49–60,73,74,77–80,93 As shown in Figure 3, the HR of the American group is 0.49, the HR of the European group is 0.47, HR of the Asian group is 0.73 and the HR of the Australian group is 0.51; MGMT promoter methylation was significantly correlated with better OS according to univariate analysis, with a combined HR of 0.50 (95% CI 0.40, 0.59).The random-effects model (the DerSimonian and Laird method) was used as significant heterogeneity was detected among these studies (p = 0.000, I2 = 50.3%).61
Thirty-one studies (six in Asia, two in America and 23 in Europe) reported the effect of MGMT promoter methylation on OS using analyses adjusted for other factors.23,26–30,33–42,45,68,75,77,82–91,93 As shown in Figure 4, the HR of the Asian group is 0.56, the HR of the American group is 0.37 and the HR of the European group is 0.44; MGMT promoter methylation was significantly correlated with better OS according to multivariate analysis, with a combined HR of 0.44 (95% CI 0.38, 0.50). The random-effects model (the DerSimonian and Laird method) was used as significant heterogeneity was detected among these studies (p = 0.000, I2 = 50.3%).61
Publication bias statistics were determined; some publication bias (Begg’s test, p<0.05) was found. Sensitivity analysis was performed to investigate the influence of a single study on the overall meta-analysis by omitting one study at a time, and the omission of any study made no significant difference, indicating that our results were statistically reliable.
Discussion
The association between the MGMT promoter methylation and GBM has been reported in many studies. Evaluations of prognostic factors, such as patients age, gender, nationality, recurrence, tumor location and excision, MGMT testing method and the style of postoperative chemoradiation for tumors are all vital to improve research pursuing new therapies for GBM. In general, the population flows more and more frequently among the continents, and most of the prognostic factors are usually determined by circumstances and nationwide medical policies. Therefore, it is more reasonable to set subgroups by areas but not by races. Our meta-analysis was performed to define the prognostic and predictive value of MGMT promoter methylation in glioblastoma patients of different continents. The major strengths of this study include the deliberate distinction of area, the relatively comprehensive sample size, the prospective data collection and the combination of the MSP and PSQ analysis to assess the MGMT promoter methylation status.
MGMT expression protects normal cells from carcinogens; however, it can also protect cancer cells from chemotherapeutic alkylating agents, which include mutations, sister chromatid exchanges, recombination and chromosomal aberrations.62 It has been shown that glial brain tumors are characterized by a low expression of MGMT, however, the activity of MGMT is commonly increased in relation to surrounding normal tissue.63,64
The data of the Adeberg et al study show that delaying postoperative chemoradiation for GBM patients – carried out in order to determine MGMT promoter status – did not have a negative impact on survival time. Indeed, initiating radiation therapy sooner than 24 days after surgery has a negative impact on progression and survival.25
In the older glioblastoma patient, MGMT promoter methylation status is still contentious on clinical decision making. For the elderly with malignant glioma, two recently published Phase III trials have evaluated the place of dose-dense/conventional temozolomide (TMZ) regimes alone as compared with conventional/hypofractionated radiotherapy.65–67 OS in methylated patients was better if TMZ treatment was applied, whereas in unmethylated patients radiotherapy alone was more effective. However, in contrast, Gutenberg et al study showed no significant differences in OS for concomitant plus adjuvant administration of TMZ, as the current standard treatment specifies, to sequentially administered TMZ.24 Concerning age, the findings of Gutenberg et al suggest that patients over 65 years of age showed significantly longer PFS and a trend toward longer OS when receiving concomitant plus adjuvant TMZ as compared to the sequential TMZ regimen. Thus, MGMT promoter methylation is an important biomarker for personalized treatment strategies in the elderly subpopulation.
It was found that GBM patients with MGMT promoter methylation had better OS and PFS than those without methylated status by univariate or multivariate analysis regardless of therapeutic intervention and area.72,74–78 MGMT gene promoter methylation levels can be used as a sensitive biomarker of using alkylating agents in GBM patients.86,88,89,92 The results suggested that MGMT promoter methylation indicated better clinical prognosis of GBM, and played an independent role with GBM development.80,82–84 Yang et al once have explored the connection between MGMT promoter methylation in glioblastoma and different race, conducting a primary conclusion, GBM patients with MGMT promoter methylation only had longer OS by multivariate analysis in Asian, but with no further exploration of subgroup.81 Also, because of population flow, it is more reasonable and accurate to set subgroup by continent but not by race. Therefore, is the prognostic significance of MGMT promoter methylation independent equally in glioblastomas of different areas?
In our univariate analysis of PFS, MGMT promoter methylation ratio of Asian groups is 0.67, the European is 0.41 and the American is 0.24. The HR of Asian groups is 0.47, the European is 0.49 and the American is 0.88. The proportion of methylation in each group was in inverse proportion to the corresponding HR. In our multivariate analysis of PFS, MGMT promoter methylation ratio of Asian groups is 0.29, the European is 0.53 and the American is 0.58. The HR of Asian groups is 0.49, the European is 0.44 and the American is 0.37. The proportion of methylation in each group was also in inverse proportion to the corresponding HR.
In our univariate analysis of OS, MGMT promoter methylation ratio of Asian groups is 0.50, the European is 0.46, the Australia is 0.36 and the American is 0.35. The HR of Asian groups is 0.73, the European is 0.47, the Australia is 0.51 and the American is 0.49. The proportion of methylation in most groups was in inverse proportion to the corresponding HR except for the Asian group. In our multivariate analysis of OS, MGMT promoter methylation ratio of Asian groups is 0.53, the European is 0.53 and the American is 0.72. The HR of Asian groups is 0.56, the European is 0.43 and the American is 0.36. The proportion of methylation in the European and American group was in inverse proportion to the corresponding HR but Asian group doesn’t follow the inverse relation.
Our meta-analysis with pooled data suggested that MGMT promoter methylation was associated with prolonged PFS in GBM patients according to both univariate analysis and multivariate analysis. From the perspective of PFS, the prognostic significance of MGMT promoter methylation is independent and basically equal in glioblastomas of different areas. Prolonged OS in GBM patients was also accompanied by MGMT promoter methylation through univariate analysis and multivariate analysis. However, from the perspective of OS, the prognostic significance of MGMT promoter methylation in the Asian group was not so important as in the European and American groups.
There are still two public questions. First, what is the most appropriate method for the assessment of methylation? The various technologies of measurement of MGMT promoter methylation sometimes show discrepant or even opposite results. It is originally regarded that MSP which evaluates the methylation status of the MGMT promoter is the best way to predict the MGMT expression of the tumor in a manner that also correlates with clinical prognosis.56 In the last 5 years, more and more studies have reported that a series of more accurate values have been obtained by PSQ compared to MSP.70 Studies with PSQ showed that this technique, having a higher reproducibility and sensitivity than MSP, is also a qualitative method. Therefore, besides MSP, our meta-analysis also absorbed measurement of MGMT promoter methylation from PSQ, which make our results more persuasive.
Second, what is the best threshold indicating methylated or unmethylated status? The definition of a prognostically relevant threshold for the percentage of MGMT methylation remains one of the most critical issues in the use of PSQ analysis. In 2015, the Receiver Operating Characteristics analysis from Villani et al showed that the best possible criteria for PSQ-detected percentage of MGMT methylation that predicted PFS and OS were 19% and 13%, respectively.23
This meta-analysis has several potential limitations that may be taken into account. First, only English and Chinese language literature studies were scanned for publication. If the search had included literature studies published in other languages, it is possible that more additional relevant trials may have been considered. Second, some ongoing studies, most of which being of high quality, were ineligible for inclusion. Therefore, limitations in quality cannot be excluded, and the pooled results of this meta-analysis may have been affected, more or less. Moreover, subgroup analysis still needs a larger number of trials to make results convincing. Additionally, we are unable to assess the effects of other clinically meaningful end points on PFS or OS, such as quality of life, patient and physician satisfaction with surgical resection and cytotoxic chemotherapy with the alkylating agent TMZ or concomitant radiotherapy, because of sparse and inconsistent reporting across studies. Finally, because all of the Asian studies included in the meta-analysis were carried out in Japan and South Korea, clinicians and pharmacists should carefully and judiciously assess the feasibility of applying the results in the clinical setting in China.
Conclusion
MGMT promoter methylation was an independent indicator of better prognosis for GBM and epigenetic MGMT gene silencing by promoter methylation associated with loss of MGMT expression may contribute to diminished DNA repair, which may be the potential mechanism that results in longer PFS and OS.71 From the perspective of PFS, the prognostic significance of MGMT promoter methylation is independent and basically equal in glioblastomas of different areas. However, from the perspective of OS, the proportion of methylation in the Asian group was not in basically inverse proportion to the corresponding HR as in European and American groups, in the univariate and multivariate analyses. The different prognosis might result from the intervention of age, percentage of MGMT methylation and the style of postoperative chemoradiation. More exploration is needed to investigate the clinical chemotherapy effect on MGMT promoter of the glioblastoma, screen a more sensitive alkylating agent combination for glioblastoma and apparent genetic targets for potential therapeutic value.
Acknowledgments
The authors thank the Intensive Care Unit of Shanghai Deji Hospital, the Ninth Clinical Medical College of Qingdao University for their help on this article. Our paper has no funding.
Author contributions
Yangyang Jiang and Wei Meng independently reviewed and extracted the data needed. Disagreements were resolved through discussion with Professor Jie Ma. All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interests in this work.
References
Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuv -ant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. | ||
Hegi ME, Liu L, Herman JG, et al. Correlation of O6-methylguanine methyl-transferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26(25):4189–4199. | ||
Verbeek B, Southgate TD, Gilham DE, Margison GP. O6-Methylguanine-DNA methyltransferase inactivation and chemotherapy. Br Med Bull. 2008;85:17–33. | ||
Gerson SL. Clinical relevance of MGMT in the treatment of cancer. J Clin Oncol. 2002;20(9):2388–2399. | ||
Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343(19):1350–1354. | ||
Bobola MS, Berger MS, Ellenbogen RG, Roberts TS, Geyer JR, Silber JR. O6-Methylguanine-DNA methyltransferase in pediatric primary brain tumors: relation to patient and tumor characteristics. Clin Cancer Res. 2001;7(3):613. | ||
Friedman HS, McLendon RE, Kerby T, et al. DNA mismatch repair and O6-alkylguanine-DNA alkyltransferase analysis and response to Temodal in newly diagnosed malignant glioma. J Clin Oncol. 1998;16(12):3851–3857. | ||
Weller M, Stupp R, Reifenberger G, et al. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol. 2009;6(1):39–51. | ||
Shilpa V, Bhagat R, Premalata CS, Pallavi VR, Ramesh G, Krishnamoorthy L. Relationship between promoter methylation & tissue expression of MGMT gene in ovarian cancer. Indian J Med Res. 2014;140(5):616–623. | ||
Felsberg J, Rapp M, Loeser S, et al. Prognostic significance of molecular markers and extent of resection in primary glioblastoma patients. Clin Cancer Res. 2009;15(21):6683–6693. | ||
Sciuscio D, Diserens D, van Dommelen D, Martinet D, Jones D, Janzer D. Extent and patterns of MGMT promoter methylation in glioblastoma- and respective glioblastoma-derived spheres. Clinical Cancer Research An Official Journal of the American Association for Cancer Research. 2011;17:255–266. | ||
Brell M, Tortosa A, Verger E, et al. Prognostic significance of O6-methylguanine-DNA methyl-transferase determined by promoter hypermethylation and immunohistochemical expression in anaplastic gliomas. Clin Cancer Res. 2005;11(14):5167–5174. | ||
Gorlia T, van den Bent MJ, Hegi ME, et al. Nomograms for predicting survival of patients with newly diagnosed glioblastoma: prognostic factor analysis of EORTC and NCIC trial 26981–22981/CE.3. Lancet Oncol. 2008;9(1):29–38. | ||
Spruance SL,Reid JE,Grace M,Samore M. Hazard ratioin clinical trials. Antimicrob Agents Chemother. 2004;48(8):2787–2792. | ||
Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into metaanalysis. Trials. 2007; 8(1):16. | ||
Cohen JF, Chalumeau M, Cohen R, Korevaar DA, Khoshnood B, Bossuyt PM. Cochran’s Q test was useful to assess heterogeneity in likelihood ratios instudies of diagnostic accuracy. J Clin Epidemiol. 2015;68(3):299–306. | ||
Deeks JJ, Altman DG. Chapter 9: analysing data and undertaking meta-analyses. Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0. 2011. Available from: http://xueshu.baidu.com/s?wd=paperuri%3A%2893155479b8a8f47eb878935af3cf9292%29&filter=sc_long_sign&tn=SE_xueshusource_2kduw22v&sc_vurl=http%3A%2F%2Fonlinelibrary.wiley.com%2Fresolve%2Freference%2FXREF%3Fid%3D10.1002%2F9780470712184.ch9&ie=utf-8&sc_us=13279804406616589552. Accessed August 29, 2017. | ||
Higgins JP, Thompson SG. Quantifying heterogeneity in a metaanalysis. Stat Med. 2002;21(11):1539–1558. | ||
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. | ||
Tang K, Jin Q, Yan W, et al. Clinical correlation of MGMT protein expression and promoter methylation in Chinese glioblastoma patients. Med Oncol. 2012;29(2):1292–1296. | ||
Motomura K, Natsume A, Kishida Y, et al. Benefits of interferon-beta and temozolomide combination therapy for newly diagnosed primary glioblastoma with the unmethylated MGMT promoter: a multicenter study. Cancer. 2011;117(8):1721–1730. | ||
Etcheverry A, Aubry M, de Tayrac M, et al. DNA methylation in glioblastoma: impact on gene expression and clinical outcome. BMC Genomics. 2010;11(1):701. | ||
Villani V, Casini B, Pace A, et al. The prognostic value of pyrosequencing-detected MGMT promoter hypermethylation in newly diagnosed patients with glioblastoma. Dis Markers. 2015;2015:604719. | ||
Gutenberg A, Bock HC, Reifenberger G, Brück W, Giese A. Toxicity and survival in primary glioblastoma patients treated with concomitant plus adjuvant temozolomide versus adjuvant temozolomide: results of a single-institution, retrospective, matched-pair analysis. Acta Neurochir (Wien). 2013;155(3):429–435. | ||
Adeberg S, Bostel T, Harrabi S, et al. Impact of delays in initiating postoperative chemoradiation while determining the MGMT promoter-methylation statuses of patients with primary glioblastoma. BMC Cancer. 2015;15:558. | ||
Kim YS, Kim SH, Cho J, et al. MGMT gene promoter methylation as a potent prognostic factor in glioblastoma treated with temozolomide-based chemoradiotherapy: a Single-Institution Study. Int J Radiat Oncol Biol Phys. 2012;84(3):661–667. | ||
Lechapt-Zalcman E, Levallet G, Dugue AE, et al. O(6)-Methylguanine-DNA methyl-transferase (MGMT) promoter methylation and low MGMT-encoded protein expression as prognostic markers in glioblastoma patients treated with biodegradable carmustine wafer implants after initial surgery followed by radiotherapy with concomitant and adjuvant temozolomide. Cancer. 2012;118(18):4545–4554. | ||
Reifenberger G, Hentschel B, Felsberg J, et al. Predictive impact of MGMT promoter methylation in glioblastoma of the elderly. Int J Cancer. 2011;131(6):1342–1350. | ||
Felsberg J, Thon N, Eigenbrod S, et al. Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas. Int J Cancer. 2011;129(3):659–670. | ||
Balana C, Carrato C, Ramirez JL, et al. Tumour and serum MGMT promoter methylation and protein expression in glioblastoma patients. Clin Transl Oncol. 2011;13(9):677–685. | ||
Lakomy R, Sana J, Hankeova S, et al. MiR-195, miR-196b, miR-181c, miR-21 expression levels and O-6-methylguanine-DNA methyltransferase methylation status are associated with clinical outcome in glioblastoma patients. Cancer Sci. 2011;102(12):2186–2190. | ||
Sonoda Y, Yokosawa M, Saito R, et al. O(6)-Methylguanine DNA methyl-transferase determined by promoter hypermethylation and immunohistochemical expression is correlated with progression-free survival in patients with glioblastoma. Int J Clin Oncol. 2010;15(4):352–358. | ||
Minniti G, Salvati M, Arcella A, et al. Correlation between O6-methylguanine-DNA methyltransferase and survival in elderly patients with glioblastoma treated with radio-therapy plus concomitant and adjuvant temozolomide. J Neurooncol. 2011;102(2):311–316. | ||
Thon N, Eigenbrod S, Grasbon-Frodl EM, et al. Predominant influence of MGMT methylation in non-resectable glioblastoma after radiotherapy plus temozolomide. J Neurol Neurosurg Psychiatry. 2011;82(4):441–446. | ||
Weller M, Felsberg J, Hartmann C, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol. 2009;27(34):5743–5750. | ||
Wemmert S, Bettscheider M, Alt S, et al. p15 promoter methylation—a novel prognostic marker in glioblastoma patients. Int J Oncol. 2009;34(6):1743–1748. | ||
Hegi ME, Diserens AC, Godard S, et al. Clinical trial substantiates the predictive value of O-6- methylguanine -DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004;10(6):1871–1874. | ||
Zawlik I, Vaccarella S, Kita D, Mittelbronn M, Franceschi S, Ohgaki H. Promoter methylation and polymorphisms of the MGMT gene in glioblastomas: a population-based study. Neuroepidemiology. 2009;32(1):21–29. | ||
Park CK, Park SH, Lee SH, et al. Methylation status of the MGMT gene promoter fails to predict the clinical outcome of glioblastoma patients treated with ACNU plus cisplatin. Neuropathology. 2009;29(4):443–449. | ||
Sonoda Y, Kumabe T, Watanabe M, et al. Long-term survivors of glioblastoma: clinical features and molecular analysis. Acta Neurochir. 2009;151(11):1349–1358. | ||
Metellus P, Coulibaly B, Nanni I, et al. Prognostic impact of O6-methylguanine-DNA methyltransferase silencing in patients with recurrent glioblastoma multiforme who undergo surgery and carmustine wafer implantation: a prospective patient cohort. Cancer. 2009;115(20):4783–4794. | ||
Cao VT, Jung TY, Jung S, et al. The correlation and prognostic significance of MGMT promoter methylation and MGMT protein in glioblastomas. Neurosurgery. 2009;65(5):866–875. | ||
Iaccarino C, Orlandi E, Ruggeri F, et al. Prognostic value of MGMT promoter status in non-resectable glioblastoma after adjuvant therapy. Clin Neurol Neurosurg. 2015;132:1–8. | ||
Barault L, Amatu A, Bleeker FE, et al. Digital PCR quantification of MGMT methylation refines prediction of clinical benefit from alkylating agents in glioblastoma and metastatic colorectal cancer. Ann Oncol. 2015;26(9):1994–1999. | ||
Shen D, Liu T, Lin Q, et al. MGMT promoter methylation correlates with an overall survival benefit in Chinese high-grade glioblastoma patients treated with radiotherapy and alkylating agent-based chemotherapy: a single-institution study. PLoS One. 2014;9(9):e107558. | ||
Melguizo C, Prados J, González B, et al. MGMT promoter methylation status and MGMT and CD133 immuno-histochemical expression as prognostic markers in glioblastoma patients treated with temozolomide plus radiotherapy. J Transl Med. 2012;10(1):250. | ||
Kanemoto M, Shirahata M, Nakauma A, et al. Prognostic prediction of glioblastoma by quantitative assessment of the methylation status of the entire MGMT promoter region. BMC Cancer. 2014;14(1):641. | ||
Christians A, Hartmann C, Benner A, et al. Prognostic value of three different methods of MGMT promoter methylation analysis in a prospective trial on newly diagnosed glioblastoma. PLoS One. 2012;7(3):e33449. | ||
Combs SE, Rieken S, Wick W, et al. Prognostic significance of IDH-1 and MGMT in patients with glioblastoma: one step forward and one step back? Radiat Oncol. 2011;6(1):115. | ||
Yang SH, Lee KS, Yang HJ, et al. O(6)-Methylguanine-DNA-methyltransferase promoter methylation assessment by microdissection-assisted methylation-specific PCR and high resolution melting analysis in patients with glioblastomas. J Neurooncol. 2012;106(2):243–250. | ||
Ellingson BM, Cloughesy TF, Pope WB, et al. Anatomic localization of O6-methylguanine DNA methyl- transferase (MGMT) promoter methylated and unmethylated tumors: a radio-graphic study in 358 de novo human glioblastomas. Neuroimage. 2012;59(2):908–916. | ||
Park CK, Kim J, Yim SY, et al. Usefulness of MS-MLPA for detection of MGMT promoter methylation in the evaluation of pseudoprogression in glioblastoma patients. Neuro Oncol. 2011;13(2):195–202. | ||
Costa BM, Caeiro C, Guimaraes I, et al. Prognostic value of MGMT promoter methylation in glioblastoma patients treated with temozolomide-based chemoradiation: a Portuguese multicentre study. Oncol Rep. 2010;23(6):1655–1662. | ||
Brandes AA, Franceschi E, Tosoni A, et al. O(6)-Methylguanine DNA-methyltransferase methylation status can change between first surgery for newly diagnosed glioblastoma and second surgery for recurrence: clinical implications. Neuro Oncol. 2010;12(3):283–288. | ||
Morandi L, Franceschi E, de Biase D, et al. Promoter methylation analysis of O6-methylguanine-DNA methyltransferase in glioblastoma: detection by locked nucleic acid based quantitative PCR using an imprinted gene (SNURF) as a reference. BMC Cancer. 2010; 10(1):48. | ||
Rivera AL, Pelloski CE, Gilbert MR, et al. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro Oncol. 2010;12(2):116–121. | ||
Zunarelli E, Bigiani N, Sartori G, Migaldi M, Sgambato A, Maiorana A. INI1 immuno-histochemical expression in glioblastoma: correlation with MGMT gene promoter methylation status and patient survival. Pathology. 2011;43(1):17–23. | ||
Karayan-Tapon L, Quillien V, Guilhot J, et al. Prognostic value of O6-methyl-guanine-DNA methyl -transferase status in glioblastoma patients, assessed by five different methods. J Neurooncol. 2010;97(3):311–322. | ||
Brandes AA, Franceschi E, Tosoni A, et al. Temozolomide concomitant and adjuvant to radiotherapy in elderly patients with glioblastoma: correlation with MGMT promoter methylation status. Cancer. 2009;115(15):3512–3518. | ||
Smith KA, Ashby LS, Gonzalez LF, et al. Prospective trial of gross-total resection with Gliadel wafers followed by early postoperative Gamma Knife radiosurgery and conformal fractionated radiotherapy as the initial treatment for patients with radiographically suspected, newly diagnosed glioblastoma multiforme. J Neurosurg. 2008;109(suppl):106–117. | ||
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. | ||
Quillien V, Lavenu A, Sanson M, et al. Outcome-based determination of optimal pyrosequencing assay for MGMT methylation detection in glioblastoma patients. J Neurooncol. 2014;116(3):487–496. | ||
Citron M, Decker R, Chen S, et al. O6-Methylguanine-DNA methyltransferase in human normal and tumor tissue from brain, lung, and ovary. Cancer Res. 1991;51(16):4131–4134. | ||
Silber JR, Mueller BA, Ewers TG, Berger MS. Comparison of O6-methyl-guanine-DNA methyltransferase activity in brain tumors and adjacent normal brain. Cancer Res. 1993;53(14):3416–3420. | ||
Thon N, Kreth S, Kreth FW. Personalized treatment strategies in glioblastoma: MGMT promoter methylation status. Onco Targets Ther. 2013;6:1363–1372. | ||
Wick W, Platten M, Meisner C, et al. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012;13(7):707–715. | ||
Malmström A, Grønberg BH, Marosi C, et al. Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trial. Lancet Oncol. 2012;13(9):916–926. | ||
Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. | ||
Gersonet SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer. 2004;4(4):296–307. | ||
Zhao H, Wang S, Song C, Zha Y, Li L. The prognostic value of MGMT promoter status by pyrosequencing assay for glioblastoma patients’ survival: a meta-analysis. World J of Surg Oncol. 2016;14:261. | ||
Chakravarti A, Erkkinen MG, Nestler U, et al. Temozolomide-mediated radiation enhancement in glioblastoma: a report on underlying mechanisms. Clin Cancer Res. 2006;12(15):4738–4746. | ||
Fiano V, Trevisan M, Trevisan E, et al. MGMT promoter methylation in plasma of glioma patients receiving temozolomide. J Neurooncol. 2014;117(2):347–357. | ||
McDonald KL, Rapkins RW, Olivier J, et al. The T genotype of the MGMT C[T (rs16906252) enhancer single-nucleotide polymorphism (SNP) is associated with promoter methylation and longer survival in glioblastoma patients. Eur J Cancer. 2013;49(2):360–368. | ||
Chen C, Huang R, MacLean A, et al. Recurrent high-grade glioma treated with bevacizumab: prognostic value of MGMT methylation, EGFR status and pretreatment MRI in determining response and survival. J Neurooncol. 2013;115(2):267–276. | ||
Ochsenbein AF, Schubert AD, Vassella E, Mariani L. Quantitative analysis of O6-methylguanine DNA methyltransferase (MGMT) promoter methylation in patients with low-grade gliomas. J Neurooncol. 2011; 103(2):343–351. | ||
Stupp R, Hegi ME, Gorlia T et al. Cilengitide combined with standard treatment for patients with newly diagnosed glioblastoma with methylated MGMT promoter (CENTRIC EORTC 26071-22072 study): a multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15(10):1100–1108. | ||
Glas M, Happold C, Rieger J, et al. Long-term survival of patients with glioblastoma treated with radiotherapy and lomustine plus temozolomide. J Clin Oncol. 2009;27(8):1257–1261. | ||
Prados MD, Chang SM, Butowski N, et al. Phase II study of erlotinib plus temozolomide during and after radiation therapy in patients with newly diagnosed glioblastoma multiforme or gliosarcoma. J Clin Oncol. 2009;27(4):579–584. | ||
Dunn J, Baborie A, Alam F, et al. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br J Cancer. 2009;101(1):124–131. | ||
Murat A, Migliavacca E, Gorlia T, et al. Stem cell-related ‘‘self-renewal’’ signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26(18):3015–3024. | ||
Yang H, Wei D, Yang K, Tang W, Luo Y, Zhang J. The prognosis of MGMT promoter methylation in glioblastoma patients of different race:a meta-analysis. Neurochem Res. 2014;39(12):1–11. | ||
Abhinav K, Aquilina K, Gbejuade H, La M, Hopkins K, Iyer V. A pilot study of glioblastoma multiforme in elderly patients: treatments, O-6-methylguanine-DNA methyltransferase (MGMT) methylation status and survival. Clin Neurol Neurosurg. 2013;115(8):1375–1378. | ||
Skiriute D, Vaitkiene P, Saferis V, et al. MGMT, GATA6, CD81, DR4, and CASP8 gene promoter methylation in glioblastoma. BMC Cancer. 2012;12:218. | ||
Carrillo JA, Lai A, Nghiemphu PL, et al. Relationship between tumor enhancement, edema, IDH1 mutational status, MGMT promoter methylation, and survival in glioblastoma. AJNR Am J Neuroradiol. 2012;33(7):1349–1355. | ||
Ohka F, Natsume A, Motomura K, et al. Wakabayashi The global DNA methylation surrogate LINE-1 methylation is correlated with MGMT promoter methylation and is a better prognostic factor for glioma. PLoS One. 2011;6(8):e23332. | ||
Wick W, Hartmann C, Engel C, et al. NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J Clin Oncol. 2009;27(35):5874–5880. | ||
Van den Bent MJ, Dubbink HJ, Sanson M, et al. MGMT promoter methylation is prognostic but not predictive for outcome to adjuvant PCV chemotherapy in anaplastic oligodendroglial tumors: a report from EORTC Brain Tumor Group Study 26951. J Clin Oncol. 2009;27(35):5881–5886. | ||
Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. | ||
Schaich M, Kestel L, Pfirrmann M, et al. MDR1 (ABCB1) gene single nucleotide polymorphism predicts outcome of temozolomide treatment in glioblastoma patients. Ann Oncol. 2009;20(1):175–181. | ||
Gerstner ER, Yip S, Wang DL, Louis DN, Iafrate AJ, Batchelor TT. Mgmt methylation is a prognostic biomarker in elderly patients with newly diagnosed glioblastoma. Neurology. 2009;73(18):1509–1510. | ||
Eoli M, Menghi F, Bruzzone MG, et al. Methylation of O6- methylguanine DNA methyltransferase and loss of heterozygosity on 19q and/or 17p are overlapping features of secondary glioblastomas with prolonged survival. Clin Cancer Res. 2007;13(9):2606–2613. | ||
Watanabe T, Katayama Y, Komine C, et al. O6-methylguanine-DNA methyltransferase methylation and TP53 mutation in malignant astro-cytomas and their relationships with clinical course. Int J Cancer. 2005;113(4):581–587. | ||
Thon N, Thorsteinsdottir J, Eigenbrod S et al. Outcome in unresectable glioblastoma:MGMT promoter methylation makes the difference. J Neurol. 2017;264 (2):350–358. | ||
Gerstner ER, Duda DG, di Tomaso E, et al. VEGF inhibitors in the treatment of cerebral edema in patients with brain cancer. Nat Rev Clin Oncol. 2009;6(4):229–236. | ||
Cheng W, Li M, Cai J, et al. A prognostic and chromosomal instability marker, refines the predictive value of MGMT promoter methylation. J Neurooncol. 2015;122(2):303–312. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.