Back to Journals » International Journal of General Medicine » Volume 14
IRF7 as an Immune-Related Prognostic Biomarker and Associated with Tumor Microenvironment in Low-Grade Glioma
Authors Peng S, Wang G, Cheng Z, Guo Z
Received 11 June 2021
Accepted for publication 22 July 2021
Published 12 August 2021 Volume 2021:14 Pages 4381—4393
DOI https://doi.org/10.2147/IJGM.S324307
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Shijun Peng, Guangyu Wang, Zhihua Cheng, Zhilin Guo
Department of Neurosurgery, The Ninth People’s Hospital Affiliated to Shanghai Jiao Tong University Medical College, Shanghai, 200001, People’s Republic of China
Correspondence: Zhihua Cheng; Zhilin Guo Tel +86 191 2170 8712
; +86 136 0169 8294
Email [email protected]; [email protected]
Purpose: Tumor microenvironment (TME) affects the occurrence and progression of low-grade glioma (LGG). The aim of this study is to identify TME-related genes that influence prognosis in LGG patients and to explore their function and role in tumor immunity.
Patients and Methods: The TME components of LGG samples in the Cancer Genome Atlas (TCGA) database were identified by the ESTIMATE method, and differentially expressed genes (DEGs) with significant differences in immune scores and stromal scores were screened out. The core genes of DEGs were screened out by protein–protein interaction (PPI) network. Furthermore, immune-related target genes significantly correlated with prognosis were identified. Survival analysis and correlation analysis showed the correlation between target genes and clinical features and prognosis. The expression differences of target genes were verified by external database Chinese Glioma Genome Atlas (CGGA). CIBERSORT software identified the proportion of tumor-infiltrating immune cells (TICs) that were significantly related to target genes. Gene set enrichment analysis (GSEA) could enrich the main functions related to high and low expression of target genes.
Results: A total of 1567 DEGs were screened out from 529 LGG samples in the TCGA database, and 146 immune-related genes affecting prognosis were found. A total of 403 core genes were obtained from PPI network. The target gene interferon regulatory factor 7 (IRF7) was significantly associated with prognosis and clinical features of the tumor. The CGGA database verified the relationship between high and low expression groups of IRF7 and prognosis. GSEA indicated that IRF7 was mainly enriched in immune-related activities, significantly correlated with T cells CD8, macrophages M1, macrophages M2 and monocytes.
Conclusion: The IRF7 is involved in immune responses in TME of LGG, which in turn influenced tumor occurrence and progression. IRF7 can act as a potential biomarker for prognosis in patients with LGG and provide a target for tumor immunotherapy.
Keywords: interferon regulatory factor 7, tumor-infiltrating immune cells, glioma, survival analysis
Graphical Abstract:
Introduction
Low-grade gliomas (LGGs) are a group of primary brain tumors with slow progression and relatively long overall survival. It is generally considered to be mainly composed of diffuse low-grade and intermediate-grade gliomas, including astrocytomas, oligoastrocytomas and oligodendrogliomas.1,2 However, histopathological classification could not fully reflect the characteristics and clinical prognosis of LGG. In 2016, the World Health Organization (WHO) classified LGG into IDH wild-type (IDHwT) cases, IDH mutants additionally carrying 1p/19q codeletion (IDHmut-codel) and IDH mutants with euploid 1p/19q (IDHmut-non-codel) based on molecular markers. The overall survival of patients with IDHwt glioma was lower than that of other subtypes.3,4 Treatment of LGG usually includes surgery, radiotherapy and chemotherapy, but postoperative recurrence of tumor, transformed into high-grade glioma, radiation-related complications, all of which resulted in a poor prognosis. Immunotherapy, by activating the patient’s immune system to restrain tumor progression and even kill tumors, has gradually become another way of treatment and has attracted more and more attention, especially in patients who have failed to respond to conventional treatments.5 Studies have shown that the tumor microenvironment (TME) infiltrating immune cells affects tumor progression and is closely related to the prognosis.6,7
The TME is the internal microenvironment for tumor cell occurrence and progress, which can provide conditions for tumor growth. Immune cells and stromal cells are the main non-tumor components, such as macrophages and neutrophils participating in tumor-related immune responses.8 In the early stage of tumor, immune cells can be activated to inhibit tumor growth. As the tumor continues to stimulate and progress, immune and stromal components change, leading to immune tolerance, and even the formation of a microenvironment that promotes tumor progression.9,10 The primary immune cells, such as macrophages, can help tumors escape immunological surveillance and block anti-tumor immune mechanisms. Previous studies have shown that the regulation of TME through gene expression affects tumor prognosis.11–13 The TME and immune biomarkers are essential for the prognostic evaluation and treatment of LGG.
In order to explore the detailed association between the prognosis of LGG and TME, we analyzed the proportion of immune and stromal components in LGG samples from the database TCGA and their relationship with tumor prognosis. The differentially expressed genes (DEGs) were obtained by differential analysis of relative values of tumor microenvironment components. We carried out univariate Cox analysis and core gene screening analysis on the DEGs and finally obtained the target genes, which were verified by the Chinese Glioma Genome Atlas CGGA database. We also analyzed the association between target genes and clinical characteristics, and conducted enrichment analysis to find out the biological functions and signaling pathways associated with target genes. Furthermore, the correlation ratio of 22 immune cells in tumor samples was analyzed by using CIBERSORT software. The association between the target gene and immune cells was further explored.
Materials and Methods
Data Source
We downloaded transcriptome data and the corresponding clinical data of 529 LGG cases from the TCGA database (https://portal.gdc.cancer.gov/).
Immune Score, Stromal Score, and ESTIMATE Score
We use the “estimate” package by R software (version: 4.0.5) to calculate the Immune Score, Stromal Score, and ESTIMATE Score in the TME of each sample, which represents the proportion of immune and stromal components in the TME of each sample. 529 LGG samples were divided into high-score and a low-score subgroups by “survival” and “survminer” packages by R software according to the median value of scores of immune, matrix and sum. The survival difference between the two groups was compared, and the survival curve was drawn by the Kaplan–Meier method, and the Log rank test was used to determine the statistical significance. P<0.05 was considered statistically significant.
Correlation Between TME Composition and Clinical Features
The “limma” and “ggpubr” packages were used to compare the correlation between generated immune, stromal scores and each clinical characteristic, using Wilcoxon and Kruskal–Wallis tests for comparison. The significant correlation threshold of p-value set as 0.05.
DEGs Screening
The DEGs between the high-score subgroup and low-score subgroup of the Immune Score and Stromal Score were obtained by R packages limma, and the |log fold change (FC)|> 1 and false discovery rate (FDR) < 0.05 were screened to determine the final differentially expressed genes.
GO and KEGG Enrichment Analysis
In order to explore the main biological functions and signaling pathways of common differentially expressed genes, gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses by using “Clusteprofiler”, “org.hs.eg.db”, “enrichplot” and “ggplot2” packages in R, and p-value < 0.05; q-value < 0.05 sieves out significant categories.
PPI Network Construction
The protein–protein interaction (PPI) network between common DEGs can be realized by the STRING database (https://string-db.org/) and Cytoscape software (version:3.8.2), and the network is constructed by using nodes with the confidence of the interaction relationship > 0.95. By calculating the number of adjacent nodes for each gene, we select the top 30 different genes with the most adjacent nodes as the core genes of the network.
COX Regression Analysis
The list of immune-related genes was downloaded from the ImmPort database (http://www.immport.org), and further immune-related genes were screened for shared DEGs. The package survival was used to further screen the immune related differential genes which were significantly correlated with prognosis (p < 0.01). The hub gene was obtained by taking the common intersection with the core gene of PPI network. Survivor analysis of the final target gene was performed on 529 LGG samples by R package survminer and survival, and p < 0.05 was statistically significant.
Correlation Between Target Genes and Clinical Characteristics
The R-packages limma and ggpubr were used to show the difference of the target gene in each clinical feature, and the correlation between the two was compared. p < 0.05 was considered statistically significant.
Verification of Target Genes
We downloaded the gene expression levels and corresponding clinical data of gliomas from the mRNAseq_693 and mRNAseq_325 databases from CGGA (http://www.cgga.org.cn/). The combined batch correction of genes from the two databases by “limma” and “sva” packages was used to obtain the gene expression of 1018 samples. The samples with missing survival time and survival status were excluded from the corresponding clinical data of 1018 samples. Patients with low-grade glioma were selected from the samples and the difference of target gene expression were verified by “survival”, “survminer”, “limma” and “ggpubr” packages.
Gene Set Enrichment Analysis
Use gene set enrichment analysis (GSEA) software (version 4.1.0) to study the function of the target gene and related signal pathways, Nom p < 0.05 and FDR q < 0.05 gene sets were considered to be significant.
Tumor Infiltrates Immune Cell Profile
To assess whether the infiltrating immune cells (TICs) in tumor samples are associated with the target gene, we used the CIBERSORT (https://cibersort.stanford.edu/) to estimate the relative abundance of 22 types of infiltrating immune cells. P < 0. 05 indicates that the estimate is accurate.
Results
Scores Correlated with Survival of LGG Patients
Survival analysis of Immune Score, Stromal Score, and ESTIMATE Score was performed by Kaplan–Meier survival analysis, respectively. Higher immune scores and stromal scores represent a higher proportion in the TME of the sample. The ESTIMATE Score is the sum of the immune and stromal scores, symbolizing the comprehensive proportion of the two components in the TME. The proportion of immune components and stromal components was significantly correlated with the prognosis of 529 cases with LGG (Figure 1A and B), and the ESTIMATE Score were also significantly associated with the prognosis (Figure 1C). These survival analyses all showed that the prognosis in the low-scores group was better than that in the high-score group, indicating that the components of TME are closely related to the occurrence and progression of LGG.
Correlation Between Scores and Clinical Characteristics
The relationship between immune score, stromal score and clinical characteristics is shown in Figure 2. Immune Score, Stromal Score, and ESTIMATE Score were significantly correlated with tumor grade respectively (P < 0.05), and the score was positively correlated with tumor grade. In addition, the score was significantly correlated with the LGG histological type (P < 0.05), and the score decreased with the decrease of astrocytoma content. All these results indicated that the composition ratio of immune score and stromal score was closely related to clinical characteristics, such as clinical grade and histological type, and affected the prognosis of patients.
The Differentially Expressed Genes
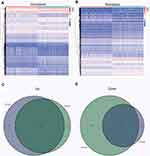
According to the median score of immune and stromal score, 529 LGG samples were divided into two groups: high-score samples and low-score samples. For immune score, between high and low groups, 1261 genes were up-regulated and 1021 genes were down-regulated. As for stromal score, 1504 genes were up-regulated and 519 genes were down-regulated in the comparison between high and low groups. All differentially expressed genes in the samples of the high and low scores of immune scores and stromal scores are displayed by heatmap (Figure 3A and B). There were a total 1567 shared DEGs that were significantly correlated in both immune score and stromal score (Figure 3C and D).
Enrichment Analyses of DEGs
The GO enrichment analysis showed the common DEGs to mainly be enriched in regulation of immune effector process, positive regulation of cell activation, positive regulation of leukocyte activation, T cell activation, adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains and T cell activation (Figure 4A and B). Similarly, the KEGG enrichment analysis also displayed enriched in Cytokine−cytokine receptor interaction, Viral protein interaction with cytokine and cytokine receptor, Chemokine signaling pathway, Human T-cell leukemia virus 1 infection (Figure 4C and D).
 |
Figure 4 Enrichment analysis of GO and KEGG in barplot, circos plots. (A and B) GO enrichment analysis for 1567 shared DEGs. (C and D) KEGG enrichment analysis for DEGs. |
Hub Gene Identification
We built a PPI network based on the STRING database and Cytoscape software (Figure 5A). There were always 403 genes expressed, among which the top 30 genes with the most adjacent nodes were selected (Figure 5B). Among the immune-related prognostic genes screened by Univariate Cox analysis (P < 0.01), the most significant 30 genes were presented (Figure 5C). The top 30 core genes in the PPI network were intersected with the most significant 30 immune-prognostic genes to obtain the hub genes. The target gene IRF7 was selected (Figure 5D).
Association of IRF7 Expressions with Survival Analysis and Clinical Features
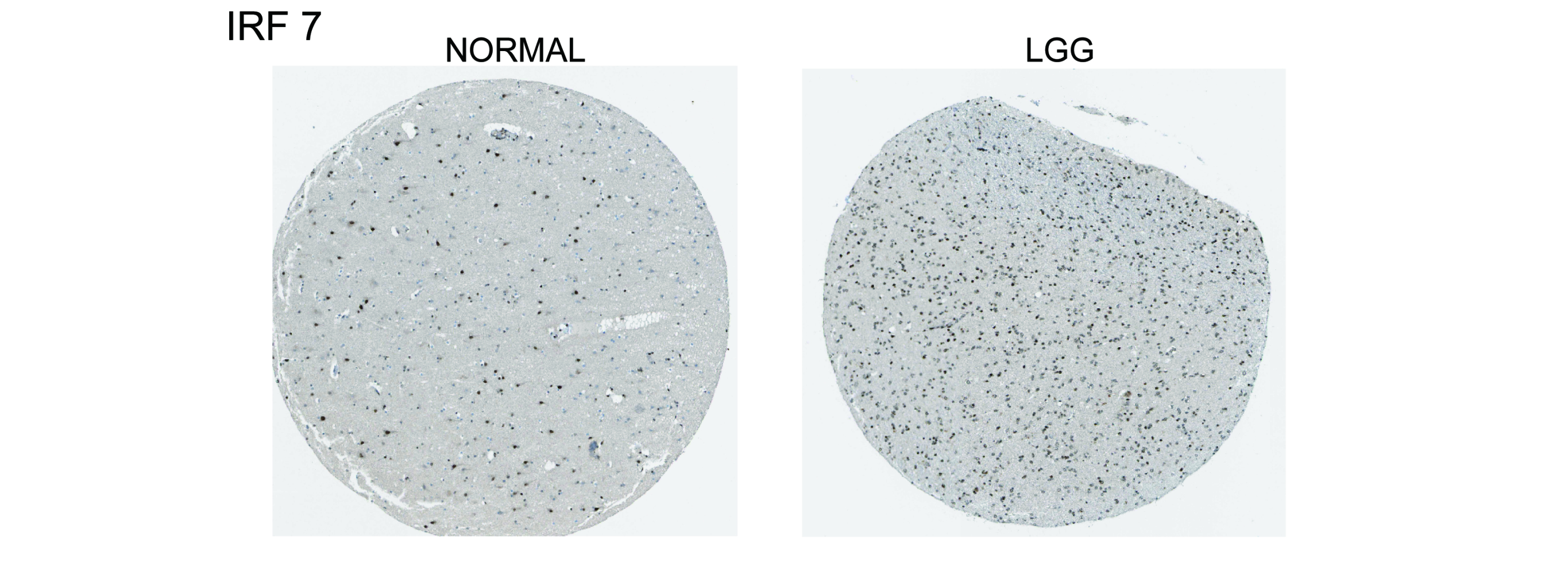
According to the median expression value of the target gene IRF7, 529 LGG samples were divided into high expression and low expression. The survival analysis showed that the IRF7 and prognosis of patients LGG was significantly correlated (p < 0.05). The expression of IRF7 in TME was negatively correlated with the prognosis of LGG patients. Patients with low expression of IRF7 had longer survival time than those with high expression (Figure 6A). Additionally, the study also showed that there were significant differences in IRF7 among different tumor grades and histological type (p < 0.05), the expression of IRF7 increased with the progression of clinical grade (Figure 6B).
Verification of Differences in IRF7 Prognosis
458 samples of LGG patients were screened from the CGGA database, and the samples were divided into the high expression group and the low expression group according to the median gene expression of the target gene IRF7 in the samples. The results also showed that the prognosis of patients in the low expression group of IRF7 was better than that in the high expression group (Figure 6C). It was also verified that the high expression of IRF7 was mainly concentrated in patients with pathological grade III (Figure 6D).
GSEA and IRF7 Expression
GSEA showed that the high IRF7 expression group was mainly enriched in antigen processing and presentation, cell adhesion molecules, the cytokine–cytokine receptor interaction, JAK/Stat signaling pathway and other tumor antigen recognition pathways (Figure 7A). Similarly, the phenotype of IRF7 is mainly enriched in the immunoreactive-related pathways, which are involved in the immune regulation of tumors (Figure 7B).
 |
Figure 7 GSEA of samples with the IRF7 high expression group. (A) The enriched gene sets in KEGG pathway. (B) The enriched gene sets in Hallmark in LGG samples. |
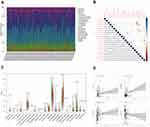
Correlation Between IRF7 and TICs
We further confirmed the relationship between target gene IRF7 expression and TICs using the CIBERSORT method. The proportion of 22 immune cell profiles in the high-expression and low-expression groups was shown (Figure 8A). The correlation between each immune cell profile was also demonstrated (Figure 8B). Differential expression analysis of IRF7 expression and 22 immune cell profiles and their correlation analysis were performed to obtain differential immune cells (Figure 8C and D). In differentially infiltrated immune cells, 4 kinds of TICs were significantly correlated with the expression of IRF7 gene, T cells CD8, macrophages M1, macrophages M2 and IRF7 were positively correlated, while monocytes and IRF7 were negatively correlated. These results further support that the expression of IRF7 affects the immune response of TME in LGG patients.
Discussion
In this study, we identified immune-related genes with different immune and stromal components in the TME of 529 patients with LGG, and combined with survival analysis, we obtained the target gene IRF7, which was significantly related to prognosis. We also compared and concluded that the expression of IRF7 was significantly different among different tumor grades and histological types. Further enrichment analysis and immune cell correlation analysis of the target gene IRF7 revealed that its main function was related to the immune system. These all further clarify that the target gene IRF7 affects the TME through immune response and then affects the occurrence, progression and final prognosis of patients with LGG, and can be used as a potential predictor for immunotherapy and prognosis evaluation of LGG patients.
Tumor microenvironment, especially immune components, plays an important role in the progression of tumors. Immune cells in the TME can be divided into those promoting tumor growth and those inhibiting tumor growth, including effector T cells, natural killer cells, dendritic cells, regulatory T cells (Tregs) and myeloid-derived suppressor cells (MDSCs). Effector T cells play a role in killing tumors based on the recognition of tumor antigens by dendritic cells. Macrophages are the main component of immune cell infiltration and can be classified into specific M1-like (activated) or M2-like (alternatively activated) functional status. M1 macrophages are considered powerful effector cells with high immune stimulation and tumor cytotoxicity by presenting antigens and producing toxic intermediates such as nitric oxide (NO).14 Macrophages can produce immunosuppressive cytokines IL-10 and transforming-growth factor β (TGFβ) to induce the formation of M2-like functional status modulates adaptive immunity and promotes angiogenesis and thus affects tumor proliferation and metastasis.15,16 The Tregs express Foxp3 to inhibit the response of immune cells to tumors. The MDSC stimulates angiogenesis through vascular endothelial growth factor (VEGF), induces the migration of cancer cells to endothelial cells, and promotes metastasis.17 The infiltrating immune cells show different biological behaviors under the influence of mediators released by tumor cells, ranging from recognizing tumor and inhibiting tumor progression to abnormal activation to help tumor escape immune surveillance. Immune cells regulate TME and affect tumor growth and prognosis.9 In ovarian cancer, CD8+ T cells and Foxp3+ regulatory T cells in the TME show a better prognosis.18 The basis of tumor immunotherapy has been considered to be the molecular identification of tumor antigens in the TME. Through the recognition of tumor specific antigen, antigen recognition specific T cells in TME can be monitored to inhibit tumor progression. In metastatic melanoma, tumor antigen-specific CD8+ T cells in the TME can stimulate cytokines to expand a large number of T cells in vitro and kill the tumor. Changes in the expression of LAG-3, which plays an immunomodulatory role in T cells, can lead to internal dysfunction of T cells, thus allowing the tumor to escape immunity.19,20
The target gene IRF7, which is significantly different in immune cell composition and stromal cell composition, can mediate tumor immunity by regulating the activity of type I interferon. Enrichment analysis showed that highly expressed IRF7 was mainly concentrated in antigen processing and presentation, cell adhesion molecules and primary immunodeficiency. IRF7 plays an important role in the immune response in the TME. Meanwhile, we also compared the expression difference and correlation of IRF7 among 529 LGG patients with different clinical grades and different histological types. We found that the expression of IRF7 in LGG patients with clinical grade III was significantly higher than that in grade II patients. There was a statistical difference between the two grades (P < 0.05). Similarly, different histological types also had different IRF7 expressions (P < 0.05), and there was no statistical difference in the expression of IRF7 among different genders, ages and races. In addition, LGG patients with low IRF7 expressions showed a better prognosis (P < 0.05) and a longer survival compared to patients with high IRF7 expression. All these indicate that IRF7 is involved in the immune response of TME and affects the prognosis of tumor.
The IRF7 is a multifunctional transcription factor, located on chromosome 11p15.5, mainly found in EB virus infection. IRF7 is induced by EBV latent protein (LMP-1), which leads to the expression of interferon, and then acts on the expression of target genes downstream of interferon to activate adaptive immune defense and induce T cell differentiation.21–23 The expression and activity of IRF7 are crucial to the normal physiological function and action of IFN. Abnormalities in IFNs are closely associated with autoimmune diseases and cancer. Pattern recognition receptor (PRR) induces the transcriptional process of IFNs through cellular cascade reaction, and IRF is an important molecule that transits PRR signal to activate immune cells.24,25 Together with IRF-3, IRF7 interferon gene expression and activation of normal T cells.26 Our results showed that IRF7 was significantly correlated with T cells CD8, macrophages M1, macrophages M2 and monocytes, further verifying that IRF7 plays an important role in tumor immunity. In addition to participating in the activation of immune-related cells in the immune response, IRF7 also affects tumors through other cytokines. Studies have suggested that IRF7 stimulates Jagging-Notch signal through IL6-Jak/STAT signal, and makes glioma stem cells express, and induces glioma angiogenesis through TNFα-IRF7-IL6 pathway, and promotes glioma proliferation.27 In terms of promoting angiogenesis, some studies have shown that IRF7 can induce inflammatory response through the NF-κBp65 pathway, and thereby promote the proliferation of vascular smooth muscle cells through the ATF3 pathway.28 IRF7 also plays an important role in the occurrence, development and metastasis of other tumors. IRF7 participates in IFN-β-mediated NK activity and can reduce bone metastasis of prostate cancer.29 Low expression of IRF7 can inhibit the proliferation of AML cells in human angiomyolipoma through the Rheb/mTOR pathway.30 In breast cancer, IRF7-mediated IFN-α reduces the production of bone marrow derived suppressor cells (MDSCs) and plays an important role in bone metastasis of breast cancer by activating CD4+ cells.31
Through the CIBERSORT algorithm, we obtained that IRF7 was significantly correlated with T cells CD8, macrophages M1, macrophages M2 and monocytes, which further verified the correlation between IRF7 and immune response. However, the specific mechanism by which IRF7 influences microenvironment immune-infiltrating cells and thus affects tumor progression needs to be further elaborated. IRF7 affects the prognosis of LGG, which provides a target spot for TME. Further experiments to verify the function and mechanism of IRF7 can provide more evidence for the research.
Conclusion
Through bioinformatics analysis of 529 LGG patients in the TCGA database, we obtained the immune-related target gene IRF7 in TME, which is related to prognosis, and discussed its function and involved signaling pathway. We believe that IRF7 affects the occurrence, progression and prognosis of tumors through immune activities in TME. It is a prognostic biomarker associated with TME and a potential target for immunotherapy in patients with LGG.
Abbreviations
IRF7, Interferon regulatory factor; WHO, World Health Organization; GBM, glioblastoma; LGG, low-grade glioma; TME, Tumor microenvironment; DEGs, differentially expressed genes; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein–protein interaction; GSEA, gene set enrichment analysis; TICs, tumor-infiltrating immune cells; AA, Astrocytoma; OAA, Oligoastrocytoma; ODG, Oligodendroglioma.
Data Sharing Statement
All public data access has been provided with the database address in the manuscript and there is no unpublished data.
Ethics Approval and Informed Consent
This was a retrospective medical record review study of public databases, written informed consent was waived.
Consent for Publication
All participants have agreed to publish.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro-Oncology. 2018;20(suppl_4):iv1–iv86. doi:10.1093/neuonc/noy131
2. Brat DJ, Verhaak RG, Aldape KD, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498.
3. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi:10.1007/s00401-016-1545-1
4. Ceccarelli M, Barthel FP, Malta TM, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. doi:10.1016/j.cell.2015.12.028
5. Xu S, Tang L, Li X, Fan F, Liu Z. Immunotherapy for glioma: current management and future application. Cancer Lett. 2020;476:1–12. doi:10.1016/j.canlet.2020.02.002
6. van den Bent MJ, Wefel JS, Schiff D, et al. Response assessment in neuro-oncology (a report of the RANO group): assessment of outcome in trials of diffuse low-grade gliomas. Lancet Oncol. 2011;12(6):583–593. doi:10.1016/S1470-2045(11)70057-2
7. Hottinger AF, Hegi ME, Baumert BG. Current management of low-grade gliomas. Curr Opin Neurol. 2016;29(6):782–788. doi:10.1097/WCO.0000000000000390
8. Kim J, Bae JS. Tumor-associated macrophages and neutrophils in tumor microenvironment. Mediators Inflamm. 2016;2016:6058147. doi:10.1155/2016/6058147
9. Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14(10):1014–1022. doi:10.1038/ni.2703
10. Ribas A. Adaptive immune resistance: how cancer protects from immune attack. Cancer Discov. 2015;5(9):915–919. doi:10.1158/2159-8290.CD-15-0563
11. Pottier C, Wheatherspoon A, Roncarati P, et al. The importance of the tumor microenvironment in the therapeutic management of cancer. Expert Rev Anticancer Ther. 2015;15(8):943–954. doi:10.1586/14737140.2015.1059279
12. Watnick RS. The role of the tumor microenvironment in regulating angiogenesis. Cold Spring Harb Perspect Med. 2012;2(12):a006676. doi:10.1101/cshperspect.a006676
13. Angell H, Galon J. From the immune contexture to the immunoscore: the role of prognostic and predictive immune markers in cancer. Curr Opin Immunol. 2013;25(2):261–267. doi:10.1016/j.coi.2013.03.004
14. Sica A, Larghi P, Mancino A, et al. Macrophage polarization in tumour progression. Semin Cancer Biol. 2008;18(5):349–355. doi:10.1016/j.semcancer.2008.03.004
15. Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23(11):549–555. doi:10.1016/S1471-4906(02)02302-5
16. Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25(12):677–686. doi:10.1016/j.it.2004.09.015
17. Zhou J, Nefedova Y, Lei A, Gabrilovich D. Neutrophils and PMN-MDSC: their biological role and interaction with stromal cells. Semin Immunol. 2018;35:19–28. doi:10.1016/j.smim.2017.12.004
18. Zhang S, Ke X, Zeng S, et al. Analysis of CD8+ Treg cells in patients with ovarian cancer: a possible mechanism for immune impairment. Cell Mol Immunol. 2015;12(5):580–591. doi:10.1038/cmi.2015.57
19. Zheng Y, Zha Y, Spaapen RM, et al. Egr2-dependent gene expression profiling and ChIP-Seq reveal novel biologic targets in T cell anergy. Mol Immunol. 2013;55(3–4):283–291. doi:10.1016/j.molimm.2013.03.006
20. Gros A, Robbins PF, Yao X, et al. PD-1 identifies the patient-specific CD8⁺ tumor-reactive repertoire infiltrating human tumors. J Clin Invest. 2014;124(5):2246–2259. doi:10.1172/JCI73639
21. Zhang L, Pagano JS. Interferon regulatory factor 7 is induced by Epstein-Barr virus latent membrane protein 1. J Virol. 2000;74(3):1061–1068. doi:10.1128/JVI.74.3.1061-1068.2000
22. Ning S, Hahn AM, Huye LE, Pagano JS. Interferon regulatory factor 7 regulates expression of Epstein-Barr virus latent membrane protein 1: a regulatory circuit. J Virol. 2003;77(17):9359–9368. doi:10.1128/JVI.77.17.9359-9368.2003
23. Ning S, Pagano JS, Barber GN. IRF7: activation, regulation, modification and function. Genes Immun. 2011;12(6):399–414. doi:10.1038/gene.2011.21
24. Sprooten J, Garg AD. Type I interferons and endoplasmic reticulum stress in health and disease. Int Rev Cell Mol Biol. 2020;350:63–118.
25. Barton GM, Kagan JC. A cell biological view of toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol. 2009;9(8):535–542. doi:10.1038/nri2587
26. Solis M, Goubau D, Romieu-Mourez R, Genin P, Civas A, Hiscott J. Distinct functions of IRF-3 and IRF-7 in IFN-alpha gene regulation and control of anti-tumor activity in primary macrophages. Biochem Pharmacol. 2006;72(11):1469–1476. doi:10.1016/j.bcp.2006.06.002
27. Jin X, Kim SH, Jeon HM, et al. Interferon regulatory factor 7 regulates glioma stem cells via interleukin-6 and Notch signalling. Brain. 2012;135(Pt 4):1055–1069. doi:10.1093/brain/aws028
28. Deng Y, Guo SL, Li JQ, et al. Interferon regulatory factor 7 inhibits rat vascular smooth muscle cell proliferation and inflammation in monocrotaline-induced pulmonary hypertension. Life Sci. 2021;264:118709. doi:10.1016/j.lfs.2020.118709
29. Lu R, Moore PA, Pitha PM. Stimulation of IRF-7 gene expression by tumor necrosis factor alpha: requirement for NFkappa B transcription factor and gene accessibility. J Biol Chem. 2002;277(19):16592–16598. doi:10.1074/jbc.M111440200
30. Makovski V, Jacob-Hirsch J, Gefen-Dor C, et al. Analysis of gene expression array in TSC2-deficient AML cells reveals IRF7 as a pivotal factor in the Rheb/mTOR pathway. Cell Death Dis. 2014;5(12):e1557. doi:10.1038/cddis.2014.502
31. Bidwell BN, Slaney CY, Withana NP, et al. Silencing of Irf7 pathways in breast cancer cells promotes bone metastasis through immune escape. Nat Med. 2012;18(8):1224–1231. doi:10.1038/nm.2830
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.