Back to Journals » Infection and Drug Resistance » Volume 12
Investigation of the prevalence of genes conferring resistance to carbapenems in Pseudomonas aeruginosa isolates from burn patients
Authors Khosravi AD , Taee S, Asarehzadegan Dezfuli A , Meghdadi H , Shafie F
Received 11 December 2018
Accepted for publication 26 February 2019
Published 7 May 2019 Volume 2019:12 Pages 1153—1159
DOI https://doi.org/10.2147/IDR.S197752
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Azar Dokht Khosravi,1,2 Shahab Taee,3 Aram Asarehzadegan Dezfuli,1 Hossein Meghdadi,2 Fatemeh Shafie2
1Infectious and Tropical Diseases Research Center, Health Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 2Department of Microbiology, School of Medicine, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 3Department of Biology, Faculty of Basic Sciences, Yasouj Branch, Islamic Azad University, Yasouj, Iran
Background and aim: Currently, the rate of hospital-acquired infections due to drug-resistant Pseudomonas aeruginosa strains shows an increasing trend and remains one of the principal reasons for mortalilty in burn patients. This study aimed to investigate the prevalence of genes conferring resistance to carbapenems in P. aeruginosa isolates from burn patients.
Methods: A total of 50 P. aeruginosa isolates were tested for antibiotic susceptibility and presence of multidrug-resistant (MDR) and extensively drug resistant (XDR) isolates, using phenotypic tests. Screening for genes conferring resistance to carbapenems was investigated by multiplex PCR method.
Results: Susceptibility testing demonstrated the highest resistance against amikacin, ceftazidime (n=44/88% each), and gentamicin (84%), while colistin sulfate was the most effective antibiotic. The rate of MDR and XDR isolates was revealed as 50% and 40% respectively. We detected the following carbapenemase genes: blaNDM (32%), followed by blaOXA-48 (18%), and blaBIC-1 (14%). This study revealed a high antibiotic resistance in P. aeruginosa isolates with a total of 40% and 50% MDR and XDR isolates respectively, and 70% carbapenem resistance. The prevalence of carbapenem conferring genes tested among carbapenem-resistant isolates was demonstrated as 65.7%.
Conclusion: Due to the prevalence of P. aeroginosa strains carrying blaOXA-48 and blaNDM genes in our hospital, more attention and implementation of effective control measures against nosocomial infection are recommended.
Keywords: Pseudomonas aeruginosa, carbapenems, encoding genes, antibiotics, drug susceptibility test
Introduction
Burn injuries is a common global public health problem, accounting for an estimated 180,000 deaths annually.1 Pseudomonas aeruginosa is an important pathogen causing a wide range of acute and chronic infections in burn patients.2 This microorganism is found in approximately 33% of all burn wounds and in 59% of extensive burns.3 Treatment and control of severe infections caused by P. aeruginosa are frequently complicated due to the limited susceptibility to antimicrobial agents and the emergence of antibiotic resistance during therapy. In 2011, the European Centre for Disease Prevention and Control and the Centers for Disease Control and Prevention redefined conventionally-acquired antibiotic resistance profiles including multidrug-resistance, extensive drug resistance, and pan-drug resistance (PDR) for some bacterial species, including P. aeruginosa.4 Literally, for P. aeruginosa specifically, multidrug-resistance means the isolate which is non-susceptible to at least one agent in ≥3 antimicrobial categories including aminoglycosides, carbapenems, and fluoroquinolones; extensive drug resistance means the isolate which is non-susceptible to at least one agent in all but two or fewer antimicrobial categories; PDR means resistant to all antibiotic classes available for empirical treatment.5 Carbapenems are the drugs used to treatmultidrug-resistant (MDR) isolates, however, the increasing frequency of carbapenem-resistant P. aeruginosa has recently been mentioned in several studies.4,6 This resistance is mediated by carbapenem-hydrolyzing enzymes, including Ambler class A (eg, KPC), B (eg, IMP, VIM), and D (eg, OXA) beta-lactamases. Epidemiological studies showed that 88.3% of MDR P. aeruginosa isolates are resistant to carbapenems, aminoglycosides, and fluoroquinolones.7 This study was conducted to investigate the prevalence of genes conferring resistance to carbapenems in P. aeruginosa isolates from burn patients, using multiplex PCR.
Materials and methods
The present study was conducted in Taleghani burn referral Hospital in Ahvaz, Iran, from March to August 2015. A total of 50 isolates of P. aeruginosa were collected from individual burn wound samples of admitted patients. The study was approved by the Institutional Review Board (IRB) and Ethics committee of the Islamic Azad University of Yasooj, after submission of the preliminary proposal, and necessary permission for sample collection was granted. Apart from this, as part of the teaching hospital's policy, referred patients were requested to sign an informed consent in case their samples were to be used for research purposes apart from routine clinical investigation. We confirm that our study was conducted in accordance with the Declaration of Helsinki.
The isolates were identified as P. aeruginosa by standard culture and biochemical tests.8 Antimicrobial susceptibility testing (AST) was performed using the agar disk diffusion method (Kirby-Bauer) on Mueller-Hinton agar (EMD Millipore, Billerica, MA, USA) plates according to the Clinical and Laboratory Standards Institute (CLSI) guideline.9 The following antimicrobial discs were used: imipenem (10 μg), meropenem (10 μg), amikacin (30 μg), gentamicin (10 μg), ciprofloxacin (5 μg), cefepime (30 μg), ceftazidime (30 μg), piperacillin (100 μg), piperacillin-tazobactam (100/10 μg), colistin-sulfate (10 μg), and aztreonam (30 μg), (MAST Diagnostics, Merseyside, UK). P. aeruginosa ATCC 27853 was used for quality control.
DNA was extracted from colonies of P. aeruginosa isolates by the simple boiling method as described elsewhere,10 and the concentration of extracted DNA was determined at 260 nm, using Nanodrop instrument (ThermoFisher Scientific, Waltham, MA, USA). Multiplex PCR was performed using previously described oligonucleotide primers to detect blaOXA-48, blaNDM, blaKPC, and blaBIC beta-lactamase genes,11 as shown in Table 1. PCR mixture was prepared in a final volume of 25 μL comprising 10× PCR buffer, 1.5 mmol of MgCl2, 0.2 mmol of each dNTPs, 5 U/μL of Taq DNA polymerase, 1 μmol of each primer, 8 μL of distilled water, and 8 ng of template DNA. The amplification was carried out in a thermocycler (Eppendorf, Germany) with the following cycling conditions: initiation denaturation at 95°C for 10 minutes, and 36 cycles of denaturation at 95°C for 30 seconds, annealing at 52°C for 40 seconds, extension at 72°C for 50 seconds, and a final extension at 72°C for 5 minutes. DNA fragments were analyzed by electrophoresis on a 2% agarose gel containing 0.5 μg/mL ethidium bromide. The bands were visualized under UV light using a gel documentation system (Protein Simple, USA). The PCR products were sent to Bioneer Corporation, Daejeon, South Korea, for sequencing.
 | Table 1 Sequence of primers used in the present study |
Results and discussion
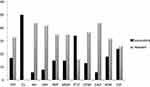
In the present study, 50 confirmed P. aeruginosa isolates were selected for further investigation. The isolates were recovered from burn wound infections of 22 (44%) male and 28 (56%) female patients with a mean age of 38.3 years. According to AST, high antibiotic resistance against most of the tested antibiotics was demonstrated in this study (40% and 50% MDR and extensively drug resistant [XDR] isolates respectively), except for four isolates (8%) that were susceptible to all applied antibiotics. The antibiotic susceptibility profile of P. aeruginosa isolates is presented in Figure 1. The highest resistance was seen against amikacin and ceftazidime (n=44/88% each) and gentamicin (84%), and the most effective antibiotic against the isolates was colistin sulfate (100%). Twenty-five isolates (50%) were resistant to at least three different classes of antibiotics according to previously described criteria,5 and were considered as MDR isolates. Moreover, according to the same criteria, 20 isolates (40%) were resistant to at least six different classes of antibiotics and were considered as XDR isolates. In Figure 2 the rate of antimicrobial susceptibility of MDR and XDR P. aeruginosa isolates to eleven antimicrobial agents is presented. The significantly high rate of 70% resistance to carbapenems (imipenem and meropenem) among the isolates may be related to frequent use of carbapenems as drug of choice for the treatment of MDR P. aeruginosa in our burn hospital for all patients, which facilitates increasing carbapenem-resistant isolates, as stated by other investigators.12 In fact, the resistance to carbapenems in the region of study started long ago, as shown in a report by Khosravi et al.13 During the past 10 years the rate of resistance to both imipenem and meropenem has increased from 41% in 2008 to 70% currently. This increase in carbapenem resistance in the same burn center, which is also true for other categories of antibiotics, could be due to several reasons, including the use of different antibiotic regimes, antibiotic overuse, presence of different persistent strains in the hospital, the quality of hygiene, and duration of hospital stays for patients with antibiotic-resistant infections.14 The high resistance (up to 90%) to carbapenems among burn patients was also reported in a recent study from Iran.15 Moreover, according to recent reports, MDR P. aeruginosa strains and resistance to carbapenems are a matter of concern worldwide as well.10 Even though carbapenems are still being used in Iran and some other countries as the last antibiotic of choice for the treatment of MDR P. aeruginosa,11 the acquisition of new resistance determinants such as extensive drug resistance remains a therapeutic challenge as effective antimicrobial therapy is severely limited.5 In the present study, 40% of P. aeruginosa isolates exhibited an XDR phenotype. This high resistance rate is very worrying. Unfortunately, according to statistics published in previous studies, we are witnessing an increasing rate in Iran,16 and other parts of the world.17
Although the most common genes conferring resistance to carbapenems are bla IMP and bla VIM, these genes were studied in our region previously,13,18 so this work was designed to investigate other non-studied genes conferring resistance to carbapenems in the region. PCR analysis showed the presence of genes conferring resistance to carbapenems in 26 (52%) isolates, of which, in 22 isolates (84.6%) only one gene was detected. blaOXA-48 gene was detected in nine (18%) isolates, seven (14%) isolates carried blaBIC-1 gene, and 14 (28%) isolates carried blaNDM-1 gene which was the most prevalent gene in the present study. blaKPC-2 gene was not detected in any of the isolates. Of the 25 MDR isolates, five, three, and five were positive for blaNDM-1, blaBIC-1, and blaOXA-48 genes respectively, while the distribution of carbapenems genes in XDR isolates was nine, three, and four, for blaNDM-1, blaBIC-1, and blaOXA-48 genes respectively. To the best of our knowledge, this is the first report of detection of blaNDM-1 gene from P. aeruginosa in the region of study. Even in a similar recent study from Iran, no incidence of blaNDM-1 positive P. aeruginosa was reported.19 NDM-1, an Ambler class B metallo-β-lactamase, renders the bacteria resistant to almost all β-lactam antibiotics, aminoglycosides, and fluoroquinolones.20 While KPC-producing organisms have been described quite often in Iran, we did not find any blaKPC-2 positive isolates in this region.15,21
In our study, nine isolates (18%) harbored blaOXA-48, seven of them were resistant to all antibiotics except for colistin, and two isolates were susceptible to carbapenems as well. Despite the fact that blaOXA-48 gene is mainly reported in Enterobacteriaceae members, there have been recent reports of isolation of this gene in P. aeruginosa strains as well.22,23 Among carbapenem-resistant isolates (n=35), 23 isolates harbored any of the tested resistance genes (65.7%). The emergence of carbapenemases, that have been acquired via mobile elements such as transposons or plasmids, can lead to a high degree of antibiotic resistance in clinical pathogens which can be transmitted among patients in a hospital setting.24 In the current study, 12 out of 35 carbapenem-resistant P. aeruginosa isolates lacked carbapenemase genes (Table 2). It is critical to note that carbapenemases are not the only mechanisms of acquired resistance to carbapenems,25 and various resistance mechanisms have been involved in antibiotic resistance in nosocomial pathogens. This study, due to its qualitative description design, had a few limitations. Because of limited time, the current study focused on the main problem faced by the burn center, ie, antibiotic ineffectiveness. Detailed research work is needed in future on the mechanisms of antibiotics, and mainly carbapenem resistance, for a better understanding of the nature of antibiotic resistance in our setting.
 | Table 2 The distribution of encoding genes among Pseudomonas aeruginosa isolates |
Conclusion
The current study revealed the highest antibiotic resistance in P. aeruginosa isolates we have ever seen in our region, which is very alarming for our public health sector. The rate of resistance had increased to 84%–88% for some of the antibiotics used, with 50% multidrug resistance and 40% extensive drug resistance, compared to other previous studies from Iran. To clarify other resistance mechanisms apart from carbapenemases, further investigations are needed. Moreover, due to the prevalence of P. aeroginosa strains carrying blaOXA-48, blaKPC, and blaNDM genes in our hospital, greater attention and implementation of effective control measures against nosocomial infection are recommended.
Acknowledgments
This work is part of an MSc thesis by Shahab Taee,26 which was approved by the Islamic Azad University, Yasouj Branch, Yasouj, Iran, and was financially self-supported. We are grateful to the School of Medicine, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran, for supporting the work carried out in the Department of Microbiology.
Disclosure
The authors report no conflicts of interest in this work.
References
1.
2. Sadikot RT, Blackwell TS, Christman JW, Prince AS. Pathogen–host interactions in Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med. 2005;171(11):1209–1223. doi:10.1164/rccm.200408-1044SO
3. Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 2009;22(4):582–610. doi:10.1128/CMR.00040-09
4. Patel G, Bonomo R. “Stormy waters ahead”: global emergence of carbapenemases. Front Microbiol. 2013;4(48):1–17. doi:10.3389/fmicb.2013.00001
5. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug resistant, extensively drug resistant and pandrug resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi:10.1111/j.1469-0691.2011.03570.x
6. Codjoe FS, Donkor ES. Carbapenem resistance: a review. Med Sci. 2017;6(1):1–28.
7. Cobos-Trigueros N, Solé M, Castro P, et al. Acquisition of Pseudomonas aeruginosa and its resistance phenotypes in critically ill medical patients: role of colonization pressure and antibiotic exposure. Crit Care. 2015;19(1):218. doi:10.1186/s13054-015-0916-7
8. Mahon CR, Lehman DC, Manuselis G
9. Patel JB, Cockerill IIIFR, Eliopoulos GM, et al. Performance Standards for Antimicrobial Susceptibility Testing (M100S); 26th Ed. Clinical and Laboratory Standards Institute (CLSI). USA: Wayne; 2016.
10. Maroui I, Barguigua A, Aboulkacem A, et al. First report of VIM-2 metallo-β-lactamases producing Pseudomonas aeruginosa isolates in Morocco. J Infect Chemother. 2016;22(3):127–132. doi:10.1016/j.jiac.2015.11.008
11. Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. doi:10.1016/j.diagmicrobio.2010.12.002
12. Wolter DJ, Acquazzino D, Goering RV, Sammut P, Khalaf N, Hanson ND. Emergence of carbapenem resistance in Pseudomonas aeruginosa isolates from a patient with cystic fibrosis in the absence of carbapenem therapy. Clin Infect Dis. 2008;46(12):137–141. doi:10.1086/524080
13. Khosravi AD, Mihani F. Detection of metallo-β-lactamase–producing Pseudomonas aeruginosa strains isolated from burn patients in Ahwaz, Iran. Diagn Microbiol Infect Dis. 2008;60(1):125–128. doi:10.1016/j.diagmicrobio.2007.08.003
14. van Kleef E, Luangasanatip N, Bonten MJ, Cooper BS. Why susceptible bacteria are resistant to hospital infection control. Wellcome Open Res. 2017;2:1–32. doi:10.12688/wellcomeopenres.10308.1
15. Lari AR, Azimi L, Soroush S, Taherikalani M. Low prevalence of metallo-beta-lactamase in Pseudomonas aeruginosa isolated from a tertiary burn care center in Tehran. Int J Immunopathol Pharmacol. 2015;28(3):384–389. doi:10.1177/0394632015578343
16. Shokri D, Khorasgani MR, Zaghian S, et al. Determination of acquired resistance profiles of Pseudomonas aeruginosa isolates and characterization of an effective bacteriocin-like inhibitory substance (BLIS) against these isolates. Jundishapur J Microbiol. 2016;9(8):1–8. doi:10.5812/jjm
17. Palavutitotai N, Jitmuang A, Tongsai S, Kiratisin P, Angkasekwinai N. Epidemiology and risk factors of extensively drug-resistant Pseudomonas aeruginosa infections. PLoS One. 2018;13(2):1–13. doi:10.1371/journal.pone.0193431
18. Moosavian M, Rahimzadeh M. Molecular detection of metallo-β-lactamase genes, blaIMP-1, blaVIM-2 and blaSPM-1 in imipenem resistant Pseudomonas aeruginosa isolated from clinical specimens in teaching hospitals of Ahvaz, Iran. Iranian J Microbiol. 2015;7(1):2.
19. Tarashi S, Goudarzi H, Erfanimanesh S, Pormohammad A, Hashemi A. Phenotypic and molecular detection of metallo-beta-lactamase genes among imipenem resistant Pseudomonas aeruginosa and Acinetobacter baumannii strains isolated from patients with burn injuries. Arch Clin Infect Dis. 2016;11(4):1–6. doi:10.5812/archcid
20. Bushnell G, Mitrani-Gold F, Mundy LM. Emergence of New Delhi metallo-β-lactamase type 1-producing enterobacteriaceae and non-enterobacteriaceae: global case detection and bacterial surveillance. Int J Infect Dis. 2013;17(5):e325–333. doi: 10.5812/jjm.27249. Published online 2016 Mar 3.
21. Falahat S, Mana Shojapour M, Sadeghi A. Detection of KPC carbapenemase in Pseudomonas aeruginosa isolated from clinical samples using modified hodge test and boronic acid phenotypic methods and their comparison with the polymerase chain reaction. Jundishapur J Microbiol. 2016;9(9):e27249. doi:10.5812/jjm
22. Evans BA, Amyes SGB. OXA –lactamases. Clin Microbiol Rev. 2014;27:241–263. doi:10.1128/CMR.00117-13
23. Mohamed SE, Alobied A, Mohamed Hussien W, Saeed MI. blaOXA-48 carbapenem resistant Pseudomonas aeruginosa clinical isolates in Sudan. J Adv Microbiol. 2018;10(4):1–5. doi:10.9734/JAMB/2018/34964
24. Juan CN, Oliver A. Carbapenemases in Pseudomonas spp. Enferm Infecc Microbiol Clin. 2010;28:19–28. doi:10.1016/S0213-005X(10)70004-5
25. Savard P, Perl TM. Combating the spread of carbapenemases in Enterobacteriaceae: a battle that infection prevention should not lose. Clin Microbiol Infect. 2014;20(9):854–861. doi:10.1111/1469-0691.12748
26. Taee S. Investigation of prevalence of genes encoding resistance to carbapenems in Pseudomonas areuginosa isolates from burn patients in Taleghani burn hospital, Ahvaz, Iran [Msc thesis]. Yasouj: Faculty of Basic Sciences, Islamic Azad University; 2016.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.