Back to Journals » Drug Design, Development and Therapy » Volume 15
Investigation of the Active Ingredients and Mechanism of Hudi Enteric-Coated Capsules in DSS-Induced Ulcerative Colitis Mice Based on Network Pharmacology and Experimental Verification
Authors Ding P , Liu J, Li Q , Lu Q , Li J, Shi R, Shi L, Mao T, Ge D, Niu H, Peng G , Wang Z
Received 30 June 2021
Accepted for publication 8 September 2021
Published 8 October 2021 Volume 2021:15 Pages 4259—4273
DOI https://doi.org/10.2147/DDDT.S326029
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Anastasios Lymperopoulos
Panghua Ding,1,* Jiajing Liu,2,* Qiuyi Li,2 Qiongqiong Lu,3 Junxiang Li,3 Rui Shi,3 Lei Shi,2 Tangyou Mao,3 Dongyu Ge,4 HaiJun Niu,5 Guiying Peng,2 Zhibin Wang3
1Department of Graduate School, Beijing University of Chinese Medicine, Beijing, People’s Republic of China; 2Department of Immunology and Microbiology, School of Life Sciences, Beijing University of Chinese Medicine, Beijing, People’s Republic of China; 3Department of Gastroenterology, Dong Fang Hospital, Beijing University of Chinese Medicine, Beijing, People’s Republic of China; 4Experimental Teaching Center, School of Traditional Chinese Medicine, Beijing University of Chinese Medicine, Beijing, People’s Republic of China; 5Anhui Joyfar Pharmaceutical Research Institute Co. Ltd, Hefei, Anhui, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Guiying Peng; Zhibin Wang Email [email protected]; [email protected]
Background: Hudi enteric-coated capsule (HDC) is a Chinese medicine prescribed to treat ulcerative colitis (UC). However, its anti-inflammatory active ingredients and mechanisms remain unknown. This study aimed to investigate the active components of HDC and explore its potential mechanisms against UC by integrating network pharmacology and experimental verification.
Methods: A DSS-induced colitis murine model was established to validate the efficacy of HDC by detecting disease activity index (DAI) and histopathological changes. Network pharmacological analysis was performed to identify the active compounds and core targets of HDC for the treatment of UC. The main compounds in HDC were identified by high-performance liquid chromatography. The relative expressions of HDC’s core targets were also determined in vivo. Finally, molecular docking was applied to model the interaction between HDC and target proteins.
Results: In an in vivo experiment, HDC, especially the middle-dose HDC, effectively reduced clinical symptoms of UC, including weight loss, bloody stool, and colon shortening. Besides, the severity of colitis was considerably suppressed by HDC as evidenced by reduced DAI scores. A total of 118 active compounds and 69 candidate targets from HDC closely related to UC progression were identified via network pharmacology. Enrichment analysis revealed that the key targets of HDC correlated with the expressions of PTGS2, TNF-α, IL-6, and IL-1β. Meanwhile, these cytokines were enriched in various biological processes through the IL-17/JAK2/STAT3 signaling pathway. The middle-dose HDC contributed more to ameliorating DSS-induced colitis through this signaling pathway than other dosages. Nine components binding to JAK2, STAT3, IL-17 and IL-6 were identified by molecular docking, confirming again the inhibition effects of HDC on the IL-17/JAK2/STAT3 signaling pathway.
Conclusion: The HDC treatment, particularly the middle-dose, exerted an anti-UC effect in a multi-component, multi-target, and multi-mechanism manner, especially inhibiting the IL-17/JAK2/STAT3 signaling pathway to downregulate the secretion of proinflammatory cytokines.
Keywords: Hudi enteric-coated capsule, ulcerative colitis, network pharmacology, IL-17/JAK2/STAT3 pathway
Graphical Abstract:
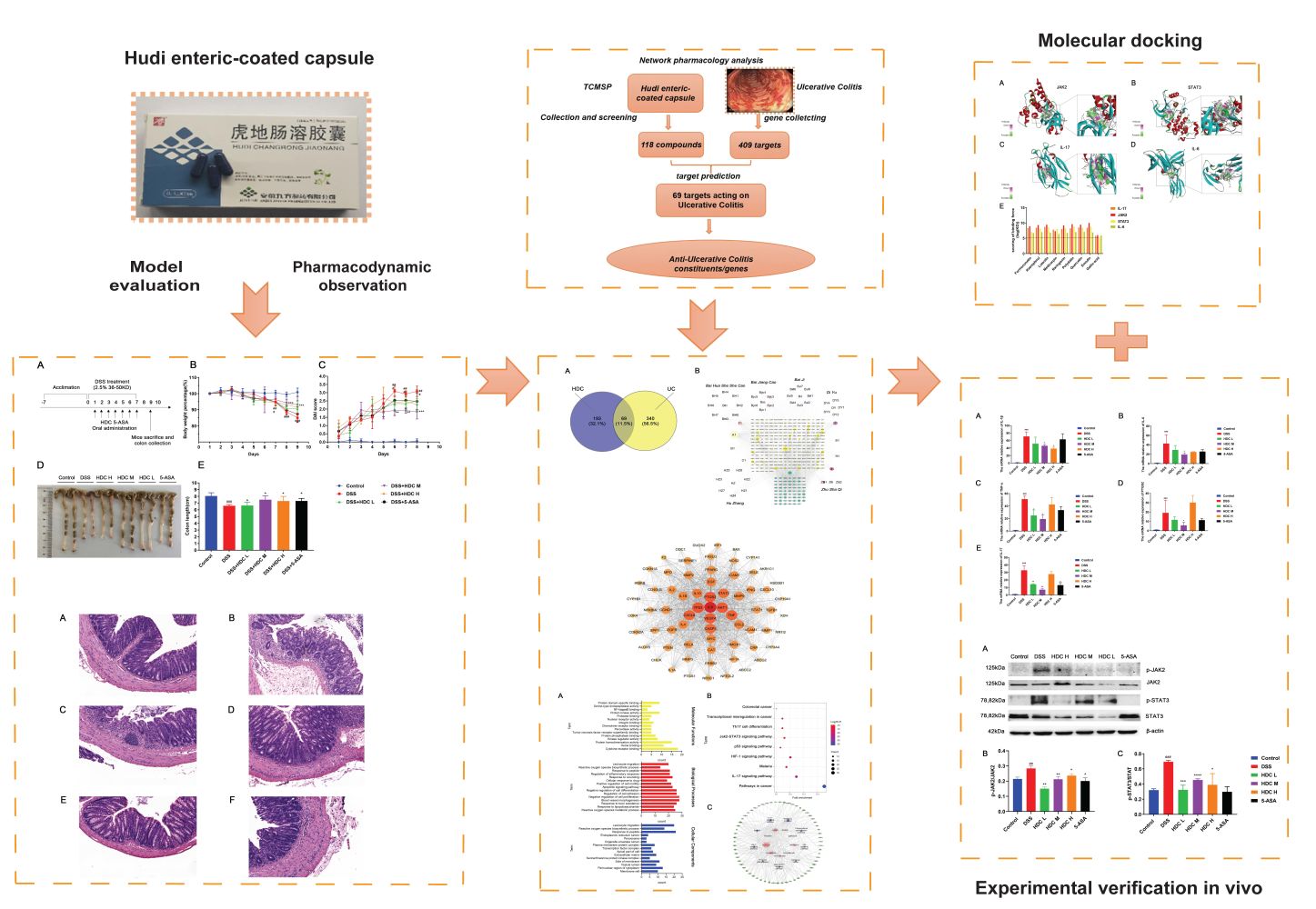
Introduction
Ulcerative colitis (UC) is one of the most common forms of inflammatory bowel disease world.1 The disease is characterized by chronic, persistent, recurrent, and nonspecific intestinal ulcer and mucosal inflammation initiating in the rectum and extending proximally in the colon in a continuous manner to varying extents.2,3 Inflammation in UC is typically limited to the mucosal layer, resulting in the occurrence of superficial damage to the bowel wall.4 The hallmark symptoms of UC include abdominal pain, chronic diarrhea, bloody stool, weight loss, as well as rectal urgency and tenesmus.5 In the past decades, the incidence of IBD has been on the rise and has become a global public health challenge.6 The pathophysiological mechanisms of UC involve the loss of epithelial barrier integrity, a dysregulated immune response, leukocyte recruitment, and the altered gut microbiota.2 To date, the first-line treatment, including 5-aminosalicylic acid, corticosteroids, immunomodulators, and biological agents (eg, adalimumab), is usually used for the treatment of UC. However, these agents are not entirely effective or can even exacerbate UC during the medication period, and are often accompanied by various side effects. Therefore, new treatment methods for UC are being investigated in various medical fields, including traditional Chinese medicine (TCM). TCM has a long history of application in the treatment of UC due to its therapeutic efficacy and few adverse events.7–10 Therefore, deciphering the pharmacological mechanism of TCM contributes to developing novel curing agents for UC.
Hudi enteric-coated capsule (HDC), a classical Chinese patent medicine, has been widely used for the treatment of mild to moderate active UC.11 HDC is composed of the following eight well-established Chinese herbs: Polygonum cillinerve (Nakai), Ohwi (Chinese name: Zhu Sha Qi), Polygonum cuspidatum Sieb. et Zucc. (Hu Zhang), Hedyotis diffusa Willd (Bai Hua She She Cao), Patrinia scabiosaefolia Fisch. ex Trev. (Bai Jiang Cao), Limonium bicolor (Bag.) Kuntze (Er Se Bu Xue Cao), Sanguisorba officinalis L. (Di Yu), Bletilla striata (Bai Ji), and Glycyrrhiza uralensis Fisch (Gan Cao) (Supplementary Table 1). HDC may exert therapeutic actions against UC via anti-inflammatory and antioxidant effects. However, the comprehensive ingredients in HDC, their multiple actions, as well as their underlying mechanisms against UC, remain unclear and need further study.
Network pharmacology is an emerging analytical tool that was initially used for drug discovery and development. It incorporates various disciplines and techniques, including genomics, proteomics, and systems biology, and provides a holistic approach for the elucidation and comprehension of drug-target relationships. This approach involves the construction of a “compound-gene-disease” interaction network and offers new strategies for designing and developing new drugs.12,13 The content and bioactivity of compounds in formulae are important factors affecting efficacy. Therefore, network pharmacology offers unique advantages and demonstrates the potential in the prediction and identification of active ingredient clusters and action targets of TCM formulations.
The HDC intervention is commonly used for the treatment of clinical UC at acute exacerbation. In this study, we, therefore, established an acute murine colitis model induced by dextran sulfate sodium (DSS) to investigate the efficacy of HDC. Next, we conducted network pharmacology based on the major components of HDC as well as compound-target-disease interactions to analyze the targets of the active ingredients in HDC and explored their mechanisms of action in vivo. Finally, we performed molecular docking experiments to validate the results of network pharmacology together with the results of animal experiment results. To our knowledge, this study is the first to thoroughly investigate the efficacy of HDC protecting against UC via network pharmacology, molecular docking techniques, and experimental verification.
Materials and Methods
Chemicals and Reagents
HDC was provided by Anhui Joyfar Pharmaceutical Co., Ltd. (200301, Bozhou, China). 5-aminosalicylic acid (5-ASA) was purchased from Losan Pharma GmbH (German). DSS was obtained from MP Biomedicals (Solon, OH, USA). A fecal occult blood test kit was obtained from the Nanjing Jiancheng Bioengineering Institute (Nanjing, China). A total RNA extraction kit, first-strand cDNA reverse transcription kit, PCR kit, and primers were obtained from Japan TaKaRa Technology (Japan). Anti-JAK2, anti-phospho-JAK2, anti-STAT3, and anti-phospho-STAT3 antibodies were purchased from Cell Signaling Technology (Buckinghamshire, UK). Reference standards, including emodin, polydatin, gallic acid, pebbles, quercetin, naringenin, alfalfa, luteolin, kaempferol, isorhamnetin, and formononetin (purity >98.0 %), were purchased from the National Institute for the Control of Pharmaceutical and Biological Products (Beijing, China) and Wuhan Tianzhi Biotechnology (Wuhan, China). Methanol and acetonitrile of high-performance liquid chromatography (HPLC) grade were purchased from Merck (Darmstadt, Germany).
Identification of Compound-Related and UC-Related Targets
First, data on all chemical components of the eight Chinese herbs in HDC were queried in the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, http://tcmspw.com/tcmsp.php).14 The active compounds in HDC were selected according to the thresholdBPs of Drug-like (DL) ≥ 0.18 and Oral bioavailability (OB) ≥ 30%. Then, targets related to the active compounds were screened from the TCMSP database, and the gene name and gene ID of the targets were obtained from the UniProt database (https://www.uniprot.org/). Data on UC-related targets were collected from the GeneCards (https://www.genecards.org/),15 OMIM (https://www.omim.org/),16 TTD (http://db.idrblab.net/ttd/), and DisGeNET (http://disgenet.org/) databases.17 After removing information on duplicate genes, data on the overlapping target genes related to UC and active compounds were retained as candidate targets.
Protein-Protein Interaction (PPI) Network Analysis
The candidate target data were inputted into String 11.0 (https://string-db.org/) for PPI analysis. The species was limited to Homo sapiens and confidence scores were limited to those ≥ 0.9.18
Gene Ontology (GO) Term Enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analyses
Metascape (https://metascape.org) was used to process data and visualize the results of GO term enrichment analysis (including biological process, cellular component, and molecular function) and KEGG pathway analysis. GO terms and KEGG pathways with P < 0.05 were considered statistically significant. We used GraphPad Prism 8.0 to generate histograms.
Molecular Docking
Data on active compounds of HDC were obtained from a component-target-pathway network and target genes were selected from the PPI network. A MOL 2 file containing information on target active compounds was obtained from the TCMSP database (http://ibts.hkbu.edu.hk/LSP/tcmsp.php). With the aid of the RCSB PDB database (https://www.rcsb.org), 3D structures were created and data were stored in PDB file format. Data on proteins and ligands were saved in the PDBQT format using Autodock Tools. After the completion of docking, compounds and target proteins with the highest docking scores and stable conformations were selected for further visualization using Pymol 2.3.2.
HPLC Analysis of HDC
We used an LC-10AT HPLC system (Shimadzu, Japan) for the chemical characterization of HDC. The contents of gallic acid, polydatin, quercetin, luteolin, naringenin, kaempferol, isorhamnetin, formononetin, and emodin were detected as markers based on the Chinese Pharmacopoeia 2015. HDC was grounded and weighed (2.0 g), 20 mL of methanol was added, and the mixture was stirred. The sample was extracted using ultrasound for 30 min, followed by centrifugation. A 10-μL aliquot of the supernatant was analyzed by conducting HPLC. An Agilent Eclipse XDB-C18 (4.6×250nm,5μm) was used for elution. The temperature of the column and autosampler was 25 °C. Samples were separated using a mobile phase consisting of A (methanol: acetonitrile,1:1) and B (0.1% KH2PO4 solution). The solvent gradients were 0.0–10 min with 7% A; 10–20 min with 7%–25% A; 20%–35 min with 25% A; 35–45 min with 25%–35% A; 45–80 min with 35%–38% A; 80–110 min with 38%–50% A; 110–120 min with 50%–67% A; and 120–140 min with 67% A. The flow rate was 1 mL/min, and the detection wavelength was 270 nm.
Experimental Animals
Male C57BL/6 mice (7–8 weeks old) were obtained from Charles River Animal and Science (Beijing, China, SYXK (JING) 2019–0013). The animals were housed in a specific pathogen-free facility at 20–25 °C and with a humidity of 40%–70%. All animal studies were approved by the Ethics Committee of Animal Experiments of Chinese Medicine (BUCM-4-2020082704-3138). Animal experiments were performed according to the guidelines of the Animal Welfare and Ethical Committee of Beijing University of Chinese Medicine.
Establishment of a Mouse Model of UC and HDC Treatment
A previous study showed some self-healing capability of DSS-induced colitis within several days.19 Hence, we conducted the continuous modeling alongside the HDC treatment to better uncover the effects of HDC with self-curing consequences excluded. The mice were randomly divided into six groups comprising 6 mice per group. Mice in the low-dose (0.5 × clinical equivalent dose), middle-dose (clinical equivalent dose), and high-dose (2 × clinical equivalent dose) groups were orally administered with 40 mg/kg, 80 mg/kg, and 160 mg/kg HDC once a day, respectively. Mice in the positive control group were orally administered with 50 mg/kg/day (clinical equivalent dose) 5-ASA, and those in the model and control group were orally administered with the same volume of sterilized distilled water. UC was induced using DSS.20 Briefly, 12.5 g DSS powder was dissolved in 500 mL of sterile water to prepare a 2.5% DSS solution (molecular weight: 36,000–50,000 Da). Except for the control group, the remaining groups received 2.5% DSS diluted in drinking water for seven days to induce UC. The DSS solution was replaced every two days. Mice were sacrificed on day 9 by cervical dislocation.
Evaluation of Disease Activity Index
The disease activity index (DAI), which reflects the degree of damage to the colon tissue, was determined based on weight loss, blood in the stool, and stool consistency. The DAI was scored continuously throughout the experiment and was calculated as the sum of scores assigned for weight loss, stool consistency, and rectal bleeding as per previously described methods.21 The DAI scoring system is shown in Table 1.
 |
Table 1 The DAI Scoring System |
Histopathological Staining
Colon tissues were collected, washed with precooled PBS, and immediately subjected to fixation in a 4% paraformaldehyde solution for 24 h. Paraffin-embedded sections (4 μm) were stained with hematoxylin and eosin (H&E) for the evaluation of colon morphology and inflammatory lesions.22
Western Blotting
The protein expressions of total JAK2, phosphorylated JAK2, STAT3, and phosphorylated STAT3 (Cell Signaling Technology, U.S.A) in colonic tissues were analyzed using Western blotting. Proteins in colon tissues of four mice in each group were extracted using the RIPA lysis buffer (Boster, Wuhan, China) and were quantified using an Enhanced BCA Protein Assay Kit (Beyotime, Shanghai, China). The proteins (20 μg from each sample) were separated on a 10% sodium dodecyl sulfate-polyacrylamide gel and transferred onto a polyvinylidene difluoride membrane. After subjection to blocking with 5% bovine serum albumin for 1 h, the membrane was incubated with primary antibodies (1:5,000) at 4 °C overnight. After conducting three washes with Tris-buffered saline containing 0.1% Tween-20, the membrane was incubated with secondary antibodies (1:5,000) at room temperature for 2 h. Finally, The GelDoc Go gel imaging system is used for exposure and development, and the Image J image analysis system is used to analyze the grayscale optical density of the target band. (Bio-Rad Company, America). All protein levels were normalized to those of β-actin.
Quantitative Reverse-Transcription Polymerase Chain Reaction (RT-qPCR)
Total RNA isolation from colon tissues was conducted using the Trizol reagent. Total RNA was reverse-transcribed using the PrimeScript™ RT Reagent Kit with gDNA Eraser. Next, RT-qPCR was carried out in a QuantStudio6 Flex system (Life Technologies, U.S.A) using miScript SYBR Green PCR Kit (Qiagen Ltd., Germany). The mRNA expressions of targeted genes, including IL-1β, IL-6, PTGS2, TNF-α, and IL-17, were calculated by the 2−ΔΔCt method with the normalization by an internal control (β-actin). Specific primers used in the current study were listed as follows: IL-1β forward (5′-CCCAACTGGTACATCAGCACCTCTC-3′) and reverse (5′-CCTGGGGAAGGCATTAGGAATAGTG-3′); IL-6 forward (5′-GGAGTCACAGAAGGAGTGGCTAAG-3′) and reverse (5′-AGTGAGGAATGTCCACAAACTGATA-3′);IL-17,forward (5′-CCTCAGACTACCTCAACCGTTCC −3′)and reverse (5′-AGGCTCCCTCTTCAGGACCAG-3′); TNF-α forward (5′-CTCTGTGAAGGGAATGGGTG-3′) and reverse (5′-GGGCTCTGAGGAGTAGACGATAAAG-3′); PTGS2 forward (5′-GAAAGCCCTCTACAGTGACATC-3′) and reverse (5′-GGTGCTCCAAGCTCTACCAT-3′); β-actin forward (5′-CGTTGACATCCGTAAAGACCTC-3′) and reverse (5′-ACAGAGTACTTGCGCTCAGGAG-3′).
Statistical Analysis
Numerical data were expressed as mean ± SEM. The Student’s t-test and one-way analysis of variance (ANOVA) followed by Bonferroni’s post-hoc tests were used for two and multiple comparisons, respectively. Statistical significance was set at P < 0.05 in all analyses. Statistical analyses were conducted using the SPSS 22.0 software.
Results
Protective Effects of Different Dosages of HDC Against DSS-Induced Colitis in Mice
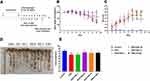
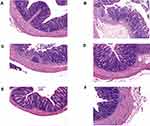
As HDC has been used widely in the clinic for the treatment of UC, we explored its in vivo therapeutic effects using a mouse model with acute UC (Figure 1A). The 2.5% DSS administration in mice for 7 days induced classical colitis symptoms as characterized by weight loss, colon shortening, and hematochezia. In the DSS group, the body weight of the mice did not decrease until day 7 (Figure 1B). As shown in Figure 1B–E, compared with the control group, mice treated with DSS presented severe pathological symptoms of UC, which mainly manifested as bloody stool, colon shortening, and weight loss. However, HDC treatments significantly increased body weight and colonic length of mice with acute colitis. The DAI values were also greater in the DSS-induced colitis group versus the control group, whereas the treatments with 40, 80, and 160 mg/kg HDC, and 5-ASA dramatically reduced the DAI scores. Moreover, severe epithelial injury, the distortion of crypts, and inflammatory cells infiltration in the mucosa and submucosa existed in the colorectal tissues of mice in the DSS group as detected by H&E staining and histopathological evaluation. HDC treatment preserved the colorectal crypt structure and ameliorated the severity of inflammation (Figure 2). Among three different dosages, HDC M was the most effective dosage against UC.
Active Ingredients of HDC and Construction of a “Drug-Target-Disease” Network
A total of 118 active compounds were identified using the TCMSP, TCMID, and Batman-TCM databases and cut-offs of OB ≥ 30% and DL ≥ 0.18. After the elimination of duplicates, we identified 272 compound-related targets, for which gene names and gene IDs were retrieved from the UniProt database. In total, 409 UC-related genes were identified in the DisGeNET, Drug Bank, and TTD databases. After merging the data on UC-related and active compound targets, 69 overlapping targets were retained as candidate targets (Figure 3A).
To elucidate the relationships among the 8 herbs present in HDC, 118 potentially active compounds, and 272 potential targets, a herb-compound-target network of HDC was constructed (Figure 3B). The network consists of 386 nodes and 1889 edges. In addition, the active ingredients of Er Se Bu Xue Cao are also observed in the other seven herbs and only in the seven herbs. Specifically, quercetin, kaempferol, luteolin, β-sitosterol, 7-methoxy-2-methylisoflavone, naringenin, formononetin, isorhamnetin, cysticercine, and medicarpin were listed in the top 10 according to the numbers of their targets by topological analysis (Supplementary Table 2), which might be considered as the major active ingredients in HDC.
PPI Network Analysis
Next, based on the 69 candidate targets, a PPI network was established by importing the gene IDs of the candidate targets into the String 11.0 database. Cytoscape 3.7.1 was used to visualize the PPI network. As shown in Figure 4, the potential compound-UC-related target network contained 69 nodes and 1203 edges. The color and size of the nodes reflect the degree value, with larger and darker red nodes indicating a greater degree centrality value. In this network, based on the connectivity degree, IL-6 (62), TP53 (60), PTGS2 (59), AKT1 (59), VEGFA (58), CXCL8 (57), TNF (57), CASP3 (55), and STAT3 (54) were identified as the top nine targets (Supplementary Table 3), which could be considered as crucial targets in the pharmacological mechanisms of HDC against UC. Notably, most potential compounds had multiple targets, corroborating the multi-compound and multi-target features of HDC.
GO and KEGG Pathway Enrichment Analysis
GO and KEGG pathway analyses were performed to understand the biological processes, molecular functions, and target-related pathways involved in the actions of HDC against UC. The 69 candidate targets were inputted into Metascape for conducting GO enrichment analysis. We found the remarkably enriched Go terms associated with targets of HDC were “reactive oxygen species metabolic process” (biological process), “membrane raft and perinuclear region of cytoplasm” (cellular component), and “cytokine receptor binding” (molecular function) (Figure 5A).
After the enrichment analysis of KEGG pathways, 97 signal pathways were obtained. According to the number of enriched genes and P value, the top 9 pathways were shown in Figure 5B. Among these, IL-17 (associated with 17 genes), JAK/STAT (associated with 11 genes), and HIF-1 signaling pathways (associated with 14 genes) were closely correlated with the pathogenesis of UC. To further explore the potential synergistic relationship of HDC on UC, the first 10 items in the enrichment pathway were used to construct the ingredient-target-pathway network (Figure 5C).
HPLC Identification of the Main Chemical Compounds in HDC
To approve the presence of those 10 ingredients screened out through network pharmacology in HDC, we chose an HPLC method for the quantitative determination of 6 monomers. In the meantime, polydatin and emodin from Reynoutria japonica Houtt and gallic acid from Sanguisorba officinalis L. were also detected by HPLC considering their high content. Figure 6A and B represented HPLC profiles of nine single standards and mixed standards, respectively. Ten batches of HDC aqueous extracts were detected. The representative profiles of HDC aqueous extracts are shown in Figure 6C and the chemical structures of the analytes and internal standards are shown in Figure 6D. The contents of gallic acid, polydatin, quercetin, luteolin, naringenin, kaempferol, isorhamnetin, formononetin, and emodin were 6.467, 4.132, 0.025, 0.136, 0.014, 0.025, 0.015, 0.035, and 4.521 (‰, w/w).
Effect of HDC on the mRNA Expressions of IL-1β, IL-17, IL-6, TNF-α, and PTGS2 in the Colon
The network pharmacological analysis suggested a few candidate targets involved in the regulation of inflammatory cytokines by HDC in UC therapy. Further, it has been reported that colon inflammation is caused by abnormal intestinal immune cell activation.23 Therefore, to validate the anti-inflammatory mechanism of HDC in UC, we evaluated the mRNA expressions of several inflammatory markers in the colon of mice with experimental UC. Compared with the control group (Figure 7), the mRNA expressions of IL-1β, IL-17, IL-6, TNF-α, and PTGS2 were markedly upregulated in the colonic tissues of DSS-treated mice (P < 0.05). However, the HDC treatments, especially the middle dose of HDC, significantly suppressed these increases (P < 0.05). These outcomes demonstrated that the middle-dose HDC treatment contributed more to inhibit the inflammatory responses in UC mice.
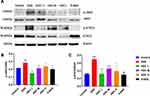
Effect of HDC on p-JAK2, JAK2, p-STAT3, and STAT3 Protein Expression in the Colon
Based on the network pharmacology analysis, the underlying mechanism of HDC in UC seemed to be strongly related to JAK2/STAT3 signaling pathway. Therefore, the protein levels of phosphorylated JAK2, JAK2, phosphorylated STAT3, and STAT3 were further detected. As shown in Figure 8, compared to the control mice, the protein expressions of p-JAK2 and p-STAT3 were significantly upregulated in the colonic tissues of DSS-treated mice. After treatment with HDC and 5-ASA, the phosphorylation of JAK2 and STAT3 protein in UC mice was significantly inhibited. These data verified that HDC prevented DSS-induced colitis via the regulation of the JAK2/STAT3 signaling pathway, as demonstrated in the network analysis.
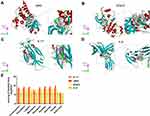
Molecular Docking Results
Network pharmacology and in vivo analyses confirmed the importance of the JAK2/STAT3 and IL-17 pathways in the exertion of effects of HDC in UC treatment. Therefore, the binding of HDC ingredients with molecules involved in these pathways was investigated using molecular docking approaches. Ingredient structures based on network pharmacology were docked with four targets, namely JAK2, STAT3, IL-17, and IL-6. The results showed that polydatin, gallic acid, quercetin, naringenin, luteolin, kaempferol, isorhamnetin, and formononetin exhibited strong affinities with JAK2, STAT3, IL-17, and IL-6 that the binding forces ranged from 6–10, 5.5–8.8, 5.8–8.9, and 5.9–7.1 log (KD), respectively (Figure 9). The comparable binding scores of the compounds might be due to their similar molecular structures, which is consistent with a previous study.24 According to the analysis based on AutoDock Tools when binding force scores are > 5.0, the tested compounds demonstrate similar molecular structures and exhibit a strong affinity for the target protein. Molecular docking results indicated that the active ingredients of HDC modulated the JAK2/STAT3 and IL-17 pathways mainly through suppressing the activities of JAK2, STAT3, and IL-17, which was in agreement with in vivo results, indicating the high reliability of the docking results.
Discussion
UC is associated with a high hospital readmission rate, and the prediction of the prognosis of UC poses great challenges. It is important to understand the immuno-inflammatory pathways involved in UC. Our study confirmed that HDC could alleviate the clinical symptoms and pathological changes in a DSS-induced mouse model with UC, which was manifested by inhibiting the DSS-induced increases in the DAI score, colon length, and weight loss. To elucidate the putative active ingredients of HDC and the mechanisms underlying its beneficial effects on UC, we conducted a comprehensive network pharmacology-based analysis. We identified 118 active compounds in HDC, 272 compound-related targets, and 409 UC-related targets from public databases. Of these, 69 targets were compound- and UC-related, thus possibly explaining the anti-UC effects of HDC. Our results indicate that HDC characteristically is multi-component and multi-target drugs with synergistic effects.
We established associations between the potential compounds and their corresponding targets by constructing a compound-target network. Based on the network pharmacology approach, we demonstrated that PTGS2 and several pro-inflammatory cytokines such as IL-1β, IL-6, and TNF-α might be potential targets for HDC compound candidates. Several studies have indicated that the upregulated mRNA expressions of PTGS2, IL-1β, IL-6, and TNF-α play important roles in the pathology of UC.25,26 TNF-α, IL-6, and IL-1β are involved in the maintenance of intestinal homeostasis and pathological processes associated with UC. Their increments are directly responsible for the infliction of mucosal injury and tissue damage and trigger disease-specific immune responses in UC.
TNF-α acts as an important molecule in intestinal inflammation. Large amounts of TNF-α drive the inflammatory process, leading to mucosal damage and the excessive secretion of other pro-inflammatory cytokines, including IL-6 and IL-1β, thus amplifying inflammation.27 Increased levels of IL-1β are also closely associated with the increased disease severity of UC.28 Stimulation with IL-1β promotes the activation and effector functions of dendritic cells, macrophages, and neutrophils.29 However, the blockade of IL-1β can ameliorate chronic intestinal inflammation in UC.30 IL-6 is another typical pro-inflammatory cytokine mainly produced by macrophages and secreted during the acute phase of inflammatory response. In UC patients, elevated levels of IL-6 have been detected in serum samples and tissue biopsies. PTGS2 (COX-2) expression is induced in response to inflammatory initiators.31,32 Many kinds of research have suggested that the inhibition of PTGS2 reduces inflammation and the severity of experimental UC by suppressing PGE2, confirming the key role in the development of UC.32,33 What’s more, COX-2-PGE2 signaling could impair intestinal epithelial regeneration and correlate with the responsiveness of TNF inhibitor in UC.34 Our results showed that the HDC administration, especially the medium dose of HDC, significantly suppressed DSS-induced inflammation by decreasing the mRNA expressions of PTGS2, TNF-α, IL-6, and IL-1β in colonic tissues of DSS-treated mice. Furthermore, PTGS2, TNF-α, IL-6, and IL-1β were identified as the potential targets of active components in HDC.
In addition, IL-17 signaling plays an important role in eliciting an inflammatory or tissue-protective response.35 It triggers inflammation by inducing the secretion of multiple cytokines and chemokines, which in turn recruit neutrophils and macrophages that contribute to tissue damage.36 Clinical patients with UC exhibit significantly greater levels of IL-17 in the serum.37 IL-17 exerts its effects by promoting tissue-resident cells to produce various matrix metalloproteinases and pro-inflammatory molecules, which act synergistically to induce the inflammatory response. Moreover, the JAK2/STAT3 signaling pathway plays a crucial part in the elicitation of responses to various cytokines and growth factors, as well as various autoimmune disorders, including UC.38–40 STAT3 and p-STAT3, controlled by the phosphorylation of JAK2, have been reported to be overexpressed in the colonic mucosa of patients with active and inactive UC.41 In our investigation, a multilevel molecular-target-disease network was constructed to systematically explore the correlations between the potential compounds of HDC and their UC targets. PPI pathways showed that IL-17 and JAK/STAT signaling pathways were involved in the therapeutic efficacy of HDC against UC. Besides, our in vivo results showed that the protein expressions of p-JAK2 and p-STAT3 were significantly increased in the colon of UC mice. Whereas, the HDC administration significantly suppressed the phosphorylation of JAK2 and STAT3. The inhibition of these two proteins might alleviate multiple pathological features such as cell growth and apoptosis by mediating the expressions of various genes in response to cell stimuli, and decelerate the degeneration of colonic inflammation.42,43 Furthermore, the mRNA level of IL-17 in colonic tissues was also remarkably arisen in UC mice while the HDC treatment restrained this enhancement. The above results suggested that HDC might inhibit the IL-17/JAK2/STAT3 signaling pathway which precludes the secretion of proinflammatory cytokines in UC mice.
Most notably, we found that IL-17, JAK/STAT, and HIF-1 signaling pathways were more closely correlated with the pathogenesis of UC than other pathways based on GO and KEGG pathway enrichment analysis. At the same time, our in vivo results demonstrated that the different dosages of HDC could inhibit the up-regulated mRNA expression of IL-17 and suppress the elevated protein expressions of p-JAK2 and p-STAT3 in the colon of DSS-treated mice. However, compared to other treatments, the middle-dose HDC was relatively more associated with the decreased weight loss and DAI scores, as well as the reduced the excessive accumulation of other inflammatory biomarkers such as IL-1β, IL-6, TNF-α, and PTGS2. Therefore, these outcomes suggested that HDC dosages could alleviate acute UC by inhibiting the IL-17/JAK2/STAT3 signaling pathway. Especially, the middle dose of HDC might be more effective on account of the greater inhibition of colonic inflammatory responses.
Finally, we adopted the molecular docking simulation to predict the active ingredients of HDC that might mediate the IL-17/JAK2/STAT3 signaling pathway. We observed that a total of 9 ingredients of HDC screened by HPLC fitted well with JAK2, STAT3, IL-17, and IL-6 at the active site pocket. Surface plasmon resonance molecular interactions and ligand binding analysis will be carried out to detect the binding affinity between these 9 ingredients and predicted target molecules in the future.
Conclusion
As summarized in our graphic abstract, by combining network pharmacology, animal experiments, and molecular docking approaches, we screened out the hub targets of HDC and explained the potential molecular mechanisms of HDC in the treatment of UC. Our results suggested that the HDC treatment, especially the middle dose of HDC, exerts a therapeutic effect on the colonic inflammation by prohibiting the expressions of pro-inflammatory cytokines such as IL-6, IL-1β, and TNF-α probably via inhibiting the IL-17/JAK2/STAT3 signaling pathway. A total of 9 active ingredients focused on the IL-17/JAK2/STAT3 signaling pathway were screened out by molecular docking. Future research is needed to explore the interaction between these active components and hub targets.
Acknowledgments
This study was supported by grants from the 13th Five-Year Plan for the National Key R&D Program of China (2018YFC1705405) and Fundamental Research Funds for Central Universities (No. 2019-JYB-TD014). We sincerely thank the Elsevier language editing service for reviewing the manuscript for language and technical content.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work. Panghua Ding and Jiajing Liu are the co-first authors, Guiying Peng and Zhibin Wang are the co-corresponding author.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569(7758):655–662. doi:10.1038/s41586-019-1237-9
2. de Souza HS, Fiocchi C. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol. 2016;13(1):13–27. doi:10.1038/nrgastro.2015.186
3. Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel JF. Ulcerative colitis. Lancet. 2017;389(10080):1756–1770. doi:10.1016/S0140-6736(16)32126-2
4. Hibi T, Ogata H. Novel pathophysiological concepts of inflammatory bowel disease. J Gastroenterol. 2006;41(1):10–16. doi:10.1007/s00535-005-1744-3
5. Halling ML, Kjeldsen J, Knudsen T, Nielsen J, Hansen LK. Patients with inflammatory bowel disease have increased risk of autoimmune and inflammatory diseases. World J Gastroenterol. 2017;23(33):6137–6146. doi:10.3748/wjg.v23.i33.6137
6. Ng SC, Shi HY, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390(10114):2769–2778. doi:10.1016/S0140-6736(17)32448-0
7. Li R, Chen Y, Shi M, et al. Gegen Qinlian decoction alleviates experimental colitis via suppressing TLR4/NF-κB signaling and enhancing antioxidant effect. Phytomedicine. 2016;23(10):1012–1020. doi:10.1016/j.phymed.2016.06.010
8. Ma Q, Ouyang Y, Meng F, et al. A review of pharmacological and clinical studies on the application of Shenling Baizhu San in treatment of ulcerative colitis. J Ethnopharmacol. 2019;244:112105. doi:10.1016/j.jep.2019.112105
9. Luo S, Wen R, Wang Q, et al. Rhubarb Peony decoction ameliorates ulcerative colitis in mice by regulating gut microbiota to restoring Th17/Treg balance. J Ethnopharmacol. 2019;231:39–49. doi:10.1016/j.jep.2018.08.033
10. Zhao Y, Luan H, Gao H, Wu X, Zhang Y, Li R. Gegen Qinlian decoction maintains colonic mucosal homeostasis in acute/chronic ulcerative colitis via bidirectionally modulating dysregulated Notch signaling. Phytomedicine. 2020;68:153182. doi:10.1016/j.phymed.2020.153182
11. Shen H, Zhu L, Hu N, et al. Hudi enteric-coated capsule combined with Mesalazine enteric-coated tablet for active ulcerative colitis: a multi-centre, randomized, double-blinded and double-simulated clinical study. Chin J Tradit West Med. 2019;39(11):1326–1331.
12. Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4(11):682–690. doi:10.1038/nchembio.118
13. Li S, Zhang B. Traditional Chinese medicine network pharmacology: theory, methodology and application. Chin J Nat Med. 2013;11(2):110–120. doi:10.3724/SP.J.1009.2013.00110
14. Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6(1):13. doi:10.1186/1758-2946-6-13
15. Stelzer G, Dalah I, Stein TI, et al. In-silico human genomics with GeneCards. Hum Genomics. 2011;5(6):709–717. doi:10.1186/1479-7364-5-6-709
16. Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: Online Mendelian Inheritance In Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43(D1):D789–798. doi:10.1093/nar/gku1205
17. Pinero J, Bravo A, Queralt-Rosinach N, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45(D1):D833–D839. doi:10.1093/nar/gkw943
18. Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(D1):D447–452. doi:10.1093/nar/gku1003
19. Liu B, Li S, Sui X, et al. Root extract of Polygonum cuspidatum Siebold & Zucc. Ameliorates DSS-induced ulcerative colitis by affecting nf-kappab signaling pathway in a mouse model via synergistic effects of polydatin, resveratrol, and emodin. Front Pharmacol. 2018;9:347
20. Ma J, Yin G, Lu Z, et al. Casticin prevents DSS induced ulcerative colitis in mice through inhibitions of NF-κB pathway and ROS signaling. Phytother Res. 2018;32(9):1770–1783. doi:10.1002/ptr.6108
21. Murthy SN, Cooper HS, Shim H, Shah RS, Ibrahim SA, Sedergran DJ. Treatment of dextran sulfate sodium-induced murine colitis by intracolonic cyclosporin. Dig Dis Sci. 1993;38(9):1722–1734. doi:10.1007/BF01303184
22. Li X, Liu Y, Wang Y, et al. Sucralose promotes colitis-associated colorectal cancer risk in a murine model along with changes in microbiota. Front Oncol. 2020;10:710. doi:10.3389/fonc.2020.00710
23. Guo T, Lin Q, Li X, et al. Octacosanol attenuates inflammation in both RAW264.7 macrophages and a mouse model of colitis. J Agric Food Chem. 2017;65(18):3647–3658. doi:10.1021/acs.jafc.6b05465
24. Liu CS, Xia T, Luo ZY, et al. Network pharmacology and pharmacokinetics integrated strategy to investigate the pharmacological mechanism of Xianglian pill on ulcerative colitis. Phytomedicine. 2021;82:153458. doi:10.1016/j.phymed.2020.153458
25. Soufli I, Toumi R, Rafa H, Touil-Boukoffa C. Overview of cytokines and nitric oxide involvement in immuno-pathogenesis of inflammatory bowel diseases. World J Gastrointest Pharmacol Ther. 2016;7(3):353–360. doi:10.4292/wjgpt.v7.i3.353
26. Katsanos KH, Papadakis KA. Inflammatory bowel disease: updates on molecular targets for biologics. Gut Liver. 2017;11(4):455–463. doi:10.5009/gnl16308
27. Rossetti S, Actis GC, Fadda M, Rizzetto M, Palmo A. The use of the anti-tumour necrosis factor monoclonal antibody—infliximab—to treat ulcerative colitis: implications and trends beyond the available data. Digest Liver Dis. 2004;36(6):426–431. doi:10.1016/S1590-8658(04)00088-X
28. Coccia M, Harrison OJ, Schiering C, et al. IL-1β mediates chronic intestinal inflammation by promoting the accumulation of IL-17A secreting innate lymphoid cells and CD4+ Th17 cells. J Exp Med. 2012;209(9):1595–1609. doi:10.1084/jem.20111453
29. Dinarello CA. Biologic basis for interleukin-1 in disease. Blood. 1996;87(6):2095–2147. doi:10.1182/blood.V87.6.2095.bloodjournal8762095
30. Simon LS. Role and regulation of cyclooxygenase-2 during inflammation. Am J Med. 1999;106(5B):37S–42S. doi:10.1016/S0002-9343(99)00115-1
31. Paiotti AP, Marchi P, Miszputen SJ, Oshima CT, Franco M, Ribeiro DA. The role of nonsteroidal antiinflammatory drugs and cyclooxygenase-2 inhibitors on experimental colitis. In Vivo. 2012;26(3):381–393.
32. Martin AR, Villegas I, Alarcón de la Lastra C. The COX-2 inhibitor, rofecoxib, ameliorates dextran sulphate sodium induced colitis in mice. Inflamm Res. 2005;54(4):145–151. doi:10.1007/s00011-004-1337-2
33. de Faria FM, Luiz-Ferreira A, Socca EA, et al. Effects of Rhizophora mangle on experimental colitis induced by TNBS in rats. Evid Based Complement Alternat Med. 2012;2012:753971. doi:10.1155/2012/753971
34. Li Y, Soendergaard C, Bergenheim FH, et al. COX-2-PGE2 signaling impairs intestinal epithelial regeneration and associates with TNF inhibitor responsiveness in ulcerative colitis. EBioMedicine. 2018;36:497–507. doi:10.1016/j.ebiom.2018.08.040
35. Majumder S, Amatya N, Revu S, et al. IL-17 metabolically reprograms activated fibroblastic reticular cells for proliferation and survival. Nat Immunol. 2019;20(5):534–545. doi:10.1038/s41590-019-0367-4
36. Singh AK, Khare P, Obaid A, et al. SUMOylation of ROR-γt inhibits IL-17 expression and inflammation via HDAC2. Nat Commun. 2018;9(1):4515. doi:10.1038/s41467-018-06924-5
37. Rampal R, Phd N, Singh A, et al. Retinoic acid is elevated in the mucosa of patients with active ulcerative colitis and displays a proinflammatory role by augmenting IL-17 and IFNγ production. Inflamm Bowel Dis. 2020;27(1):74–83.
38. Salas A, Hernandez-Rocha C, Duijvestein M, et al. JAK–STAT pathway targeting for the treatment of inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17(6):323–337. doi:10.1038/s41575-020-0273-0
39. Lin H, Honglang L, Weifeng L, Junmin C, Jiantao Y, Junjing G. The mechanism of alopolysaccharide protecting ulceralive colitis. Biomed Pharmacother. 2017;88:145–150. doi:10.1016/j.biopha.2016.11.138
40. Chu XQ, Wang J, Chen GX, Zhang GQ, Zhang DY, Cai YY. Overexpression of microRNA-495 improves the intestinal mucosal barrier function by targeting STAT3 via inhibition of the JAK/STAT3 signaling pathway in a mouse model of ulcerative colitis. Pathol Res Pract. 2018;214(1):151–162. doi:10.1016/j.prp.2017.10.003
41. Vavricka SR, Galvan JA, Dawson H, et al. Expression patterns of TNFα, MAdCAM1, and STAT3 in intestinal and skin manifestations of inflammatory bowel disease. J Crohns Colitis. 2018;12(3):347–354. doi:10.1093/ecco-jcc/jjx158
42. Yu T, Li Z, Xu L, Yang M, Zhou X. Anti-inflammation effect of Qingchang suppository in ulcerative colitis through JAK2/STAT3 signaling pathway in vitro and in vivo. J Ethnopharmacol. 2021;266:113442. doi:10.1016/j.jep.2020.113442
43. Gurung P, Dahal S, Chaudhary P, et al. Potent inhibitory effect of BJ-3105, a 6-Alkoxypyridin-3-ol derivative, on murine colitis is mediated by activating AMPK and inhibiting NOX. Int J Mol Sci. 2020;21(9):3145. doi:10.3390/ijms21093145
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.