Back to Journals » International Journal of General Medicine » Volume 14
Investigation of Potential Molecular Biomarkers for Diagnosis and Prognosis of AFP-Negative HCC
Authors Liu Z, Pu Y, Bao Y, He S
Received 8 June 2021
Accepted for publication 2 August 2021
Published 10 August 2021 Volume 2021:14 Pages 4369—4380
DOI https://doi.org/10.2147/IJGM.S323868
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Zijing Liu,1 Youwei Pu,2 Yixi Bao,2 Song He1
1Department of Gastroenterology, the Second Affiliated Hospital of Chongqing Medical University, Chongqing, 400010, People’s Republic of China; 2Department of Clinical Laboratory, the Second Affiliated Hospital of Chongqing Medical University, Chongqing, 400010, People’s Republic of China
Correspondence: Song He
Department of Gastroenterology, the Second Affiliated Hospital of Chongqing Medical University, No. 76, Linjiang Road, Chongqing, 400010, People’s Republic of China
Email [email protected]
Background: Alpha-fetoprotein (AFP) is the most important diagnostic and prognostic index of hepatocellular carcinoma (HCC). AFP-positive HCC can be easily diagnosed based on the serum AFP level and typical imaging features, but a number of HCC patients are negative (AFP < 20 ng/mL) for AFP. Therefore, it is necessary to develop novel diagnostic and prognostic biomarkers for AFP-negative HCC.
Methods: RNA data from TCGA and differential expression of lncRNAs, miRNAs, and mRNAs were downloaded to analyze the differential RNA expression patterns between AFP-negative HCC tissues and normal tissues. A lncRNA-miRNA-mRNA ceRNA regulatory network was constructed to elucidate the interaction mechanism of RNAs. Functional enrichment analysis of these DEmRNAs was performed to indirectly reveal the mechanism of action of lncRNAs. A PPI network was built using STRING, and the hub genes were identified with Cytoscape. The diagnostic value of hub genes was assessed with receiver operating characteristic (ROC) analysis. And the prognostic value of RNAs in the ceRNA was estimated with Kaplan–Meier curve analysis.
Results: A total of 131 lncRNAs, 185 miRNA, and 1309 mRNAs were found to be differentially expressed in AFP-negative HCC. A ceRNA network consisting of 12 lncRNA, 23 miRNA, and 74 mRNA was constructed. The top ten hub genes including EZH2, CCNB1, E2F1, PBK, CHAF1A, ESR1, RRM2, CCNE1, MCM4, and ATAD2 showed good diagnostic power under the ROC curve; and 2 lncRNAs (LINC00261, LINC00482), 3 miRNAs (hsa-miR-93, hsa-miR-221, hsa-miR-222), and 2 mRNAs (EGR2, LPCAT1) were found to be associated with the overall survival of AFP-negative patients.
Conclusion: This study could provide a novel insight into the molecular pathogenesis of AFP-negative HCC and reveal some candidate diagnostic and prognostic biomarkers for AFP-negative HCC.
Keywords: AFP-negative HCC, ceRNA network, diagnosis, prognosis
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignant cancers and the second leading cause of cancer-associated deaths worldwide with the 5-year survival rate of <5%.1,2 Most of the HCC cases are diagnosed at an advanced stage due to the lack of specific biomarkers and poor clinical symptoms, and over 80% of them have a poor prognosis.3 Alpha-fetoprotein (AFP) is the most important and commonly used diagnostic indicator for HCC.4,5 A previous study has indicated that AFP can promote cell growth and escape from the host immune surveillance of liver cancer cells in vivo, which is an independent risk factor associated with the prognosis.6 Only 33–65% of the tumors with a diameter of <3 cm in HCC patients had an elevated serum AFP level,1,7 and nearly 30% of the patients had normal AFP levels.8,9 Although AFP has its limitations as a biomarker for screening for liver cancer, it is still most widely used biomarker. Thus, it is crucial to identify new biomarkers for the diagnostic and prognostic evaluation of AFP-negative HCC and understand the underlying mechanisms of its tumorigenesis.
Non-coding RNA (ncRNA) is a group of noncoding RNA transcripts, which have many pivotal functions in various physiological and pathological processes.10,11 Long non-coding RNAs (lncRNAs) can regulate gene expression indirectly although they have no protein-coding function.12,13 As increasing number of studies have reported that the dysregulation of lncRNAs plays a vital role in the tumorigenesis of different types of cancer and are related to the poor prognosis in HCC,14–16 it is necessary to understand the role of lncRNAs in cancer.
The competing endogenous RNA (ceRNA) hypothesis proposed that lncRNAs can competitively bind to microRNA (miRNA) and indirectly regulate its expression levels by acting as miRNAs sponges.17–19 The ceRNA network has been shown to play a critical role in HCC pathogenesis and progression.15,20,21 However, the underlying molecular mechanisms of AFP-negative HCC and the role of ceRNA in its tumorigenesis are still unknown.
In this study, the lncRNA, miRNA, and mRNA gene expression data of HCC were downloaded from the TCGA database and screened for AFP-negative HCC. After identifying the differentially expressed RNAs (DERNAs) between AFP-negative HCC samples and adjacent non-cancerous samples, the lncRNA-miRNA-mRNA regulatory network and protein-protein interaction (PPI) network for AFP-negative HCC were constructed. Receiver operating characteristic (ROC) curves of the top 10 hub genes were used to assess the diagnostic efficiency. Meanwhile, RNAs which played a key role in the prognosis of AFP-negative HCC among the ceRNA were identified. This study will contribute to assist clinical diagnosis of liver cancer and evaluate its prognosis when the level of AFP is negative.
Materials and Methods
Data Source and Pre-Treatment
The original gene expression data and clinical information including the AFP value of HCC patients were obtained from the TCGA database (https://cancergenome.nih.gov/). The miRNA-seq data from 150 AFP-negative (AFP <20 ng/mL) HCC samples and 50 adjacent non-tumorous samples, as well as mRNA-seq data from 147 AFP-negative HCC samples and 50 adjacent non-tumorous samples were used. As data from TCGA are publicly available, no further ethical approval was required for the conduct of this study.
Differential Expression Analysis
The RNA and miRNA sequencing data were downloaded to distinguish differentially expressed lncRNAs, miRNAs, and mRNAs between the AFP-negative HCC and normal samples. After the gene expression matrix was merged, the ensemble IDs were converted to gene names. The data was calibrated and standardized, and the differential expression was identified by applying the ‘edgeR’ package of R software. The False Discovery Rate (FDR) was used to correct the P value for determining the statistical significance of multiple testing. The dysregulated RNAs were identified based on the cut-off criteria FDR <0.05 and |log2 (fold change) |>1.
CeRNA Network Construction
The lncRNA-miRNA interactions were predicted by using the miRcode (http://www.mircode.org/).22 In addition, miRDB (http://www.mirdb.org/), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw), and TargetScan (http://www.targetscan.org) were applied to predict the DEmiRNA target genes,23–25 and only miRNA-mRNA pairs found in all the three databases were utilized to construct the ceRNA networks. To further increase the reliability of the ceRNA network, the overlap between DEmiRNA target genes and the DEmRNA in AFP-negative HCC obtained by Venn diagram was used as DEmRNA. Finally, the ceRNA network based on the matched DElncRNA–DEmiRNA and DEmiRNA–DEmRNA pairs was constructed to clarify the gene interactions in AFP-negative HCC. Cytoscape software26 was used to visualize lncRNA-miRNA-mRNA network.
Functional Enrichment Analysis
To explore the potential biological processes and pathways of overlapping differentially expressed genes (DEGs), Gene Ontology (GO) enrichment at the significant level (p-value < 0.01) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses at the significant level (p-value < 0.05) was performed using Database for Annotation, Visualization and Integrated Discovery (DAVID: available online: http://david.abcc.ncifcrf.gov/).
PPI Network and Hub Genes
To explore the interrelationship of DEmRNAs, STRING (https://string-db.org/),27 a widely used interactive tool, was utilized to set up a PPI network, which was then visualized using the Cytoscape software. The top ten well connected DEmRNAs were identified as hub genes with CytoHubba. ROC curves were generated to further explore the diagnostic value of the 10 hub genes, and the area under the curve (AUC) was used to evaluate the diagnostic efficiency.
Survival Analysis
To explore the prognostic DERNA signature, differentially expressed lncRNAs, miRNAs, and mRNAs associated with survival were analyzed using the ‘Survival’ package of R software. Survival curves were estimated based on Kaplan–Meier survival curve analysis and P < 0.05 was considered as statistically significant.
Results
Differentially Expressed lncRNA, mRNA, and miRNA
A total of 131 lncRNAs (87 upregulated and 44 downregulated), 185 miRNA (141 upregulated and 44 downregulated), and 1309 mRNAs (786 upregulated and 523 downregulated) were observed to be differentially expressed in AFP-negative HCC on the criteria that |log2 (fold change)| >1, FDR <0.05. The visual volcano plots are shown in Figure 1.
Construction and Analysis of ceRNA Regulatory Network
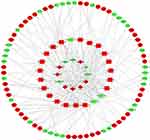
The DElncRNA-DEmiRNA and DEmiRNA-DEmRNA pairs were used to construct the ceRNA network to further understand how lncRNA mediates mRNA by binding miRNA in AFP-negative HCC. The result showed that 81 DElncRNA-DEmiRNA interactions were identified, with 12 DElncRNAs targeting 23 DEmiRNAs from the miRcode database (Table 1). The miRDB, miRTarBase, and TargetScan databases were combined to predict the candidate mRNA targets of the DEmiRNAs to improve the reliability of the results. A total of 1119 target genes that intersected with 1309 DEmRNA were obtained, resulting in 74 DEmRNA (Figure 2). Finally, the ceRNA network comprising 12 DElncRNAs, 23 DEmiRNAs, and 74 DEmRNAs was visualized by Cytoscape software (Figure 3).
 |
Table 1 12 DElncRNAs Target 23 DEmiRNAs from the miRcode Database |
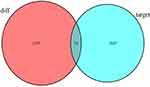 |
Figure 2 Venn diagram analysis for the intersection set between differentially expressed mRNA in AFP-negative HCC and miRNA target genes. |
Function Enrichment
To explore the function of the ceRNA network, the GO and KEGG enrichment analysis of DEmRNAs in the ceRNA network was performed using DAVID to explore the function of the ceRNA network. As shown in Figure 4, 11 GO biological process (BP) terms such as “negative regulation of apoptotic process,” “response to drug,” “positive regulation of transcription,” and so on were significantly (P < 0.01) enriched. Also, in the cellular component (CC), the significantly (P < 0.01) enriched GO terms were “nucleus,” “membrane,” “nucleoplasm,” and’ “nuclear chromatin,” and those in the molecular function (MF) were “protein kinase binding,” “chromatin binding,” “protein binding,” “ATP binding,” and “beta-catenin binding.” The KEGG results revealed that 74 DEGs were involved in 26 significantly (P < 0.05) enriched pathways (Figure 5). Among these pathways, “p53 signaling pathway”, “Hepatitis B” “PI3K-Akt” were related to hepatocarcinogenesis. And some other pathways such as “Bladder cancer,” “Colorectal cancer,” “Small cell lung cancer,” “Prostate cancer,” “MicroRNAs in cancer,” and “Pathways in cancer” were related to cancer-related pathways.
 |
Figure 4 The GO enrichment analysis of DEmRNAs in the ceRNA network of AFP-negative HCC. |
 |
Figure 5 The enriched KEGG pathway of DEmRNAs in the ceRNA network of AFP-negative HCC. |
PPI Network of DEmRNAs in the ceRNA Network
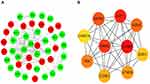
A PPI network was built to elaborate interaction relationship between the 74 DEGs using the STRING database (interaction score >0.4) and displayed using the Cytoscape software after discarding the unrelated nodes (Figure 6A). Moreover, the top ten DEmRNAs obtained by CytoHubba were identified as the hub genes including CCNB1, E2F1, RRM2, MCM4, EZH2, PBK, ATAD2, CCNE1, ESR1, CHAF1A (Figure 6B). To explore whether the top 10 hub genes have excellent diagnostic efficiency in AFP-negative HCC patients, the ROC curves based on the TCGA database were performed. The results (Figure 7) showed the AUC of CCNB1, E2F1, RRM2, MCM4, EZH2, PBK, ATAD2, CCNE1, ESR1, and CHAF1A to be 0.954, 0.949, 0.916, 0.848, 0.962, 0.929, 0.816, 0.873, 0.920, and 0.928, respectively.
Survival Analysis of RNAs in the Network
To study the prognostic capability of genes in the ceRNA network, the overall survival of HCC patients was determined with Kaplan–Meier curve analysis between DERNAs (P < 0.05 as the screening criterion). The results (Figure 8) showed that 2 lncRNAs (LINC00261, LINC00482), 3 miRNAs (hsa-miR-93, hsa-miR-221, and hsa-miR-222), and 2 mRNAs (EGR2, LPCAT1) were significantly correlated with the overall survival. High expression of LINC00482, hsa-miR-221, hsa-miR-222, and LPCAT1 was associated with poor overall survival, suggesting their potential negative role in AFP-negative HCC patients. However, high expression of LINC00261, hsa-miR-93 EGR2 was associated with good overall survival, which may be designated as AFP-negative HCC suppressor genes.
 |
Figure 8 Kaplan–Meier survival curves of lncRNAs (A and B), miRNA (C–E), mRNA (F and G) associated with overall survival in AFP-negative HCC. |
Discussion
AFP, as a conventional biomarker widely used in clinical practice, has provided critical clues for the diagnosis of HCC.28 However, there are still approximately a number of HCC patients are negative for AFP.9 In addition, the AFP level is an independent risk factor correlated with tumor differentiation, TNM stage, and survival of patients with HCC.29 Therefore, it is vital to identify more potential biomarkers for the effective diagnostic and evaluation of AFP-negative HCC.
LncRNA and miRNA, as non-coding RNA, are closely associated with the pathogenesis and progression of tumors. The emergence of lncRNA-miRNA-mRNA ceRNA revealed the regulatory relationship between non-coding and coding RNA and provided a novel guiding theory to explain the mechanism of tumorigenesis.17,30 Hence, to better understand the molecular pathogenesis involved in AFP-negative HCC, a ceRNA network was built based on the TCGA database. In this study, the DERNAs between AFP-negative HCC and normal samples were identified, and a ceRNA network comprising 12 DElncRNAs, 23 miRNAs, and 74 DEmRNAs was constructed. Among them, lncRNA FAM99B have been reported to suppress HCC progression, and the low expression of FAM99B was associated with advanced histologic grade, T-stage, and vascular invasion.31 Furthermore, lncRNA RUSC1-AS1 can promote HCC cell proliferation and inhibit HCC cell apoptosis through Notch signaling pathway, and high expression of RUSC1-AS1 indicated a poor prognosis.32 MiR-214 has been shown to be a suppressor gene in many types of cancers including HCC, and it can reduce EZH2 to suppress tumorigenesis in various types of cancer.33,34 MiR-216a and miR-137 have been demonstrated to be associated with hepatocarcinogenesis and tumor recurrence in HCC.35–37 The ceRNA network in this study identified many RNAs that play an important role in the occurrence and development of HCC.
The AFP level has been reported to be associated with the pathological grade, progression, and prognosis of HCC.38,39 Several studies have identified many prognostic indicators for HCC. However, there are few studies on the prognostic indicators of AFP-negative HCC based on a ceRNA network. Therefore, in this study, the relationship between aberrantly expressed RNAs in the ceRNA and prognosis was assessed to determine the potential prognostic indicators for AFP-negative HCC. It was noted that 2 lncRNAs (LINC00261, LINC00482), 3 miRNAs (hsa-miR-93, hsa-miR-221, and hsa-miR-222) and 2 mRNAs (EGR2, LPCAT1) were correlated with the overall survival in AFP-negative HCC patients. Among these RNAs, LINC00261 has been reported to suppress cell proliferation and invasion in HCC, and associate with AFP levels and postoperative outcome of HCC.40,41 Furthermore, the low expression of LINC00261 in AFP-negative HCC also showed a poor prognosis in the present study. Xuan L suggested LPCAT1 to be a potent target molecule for inhibiting HCC progression.42 The high expression of LPCAT1 demonstrated a poor prognosis in this study, indicative of a positive correlation with the pathologic stage of AFP-negative HCC.
To further understand the ceRNA network, GO and KEGG analyses were performed to reveal the functions and pathways of 74 DEmRNAs in the ceRNA. The GO analyses demonstrated that DEmRNAs may focus on positive regulation transcription, DNA-templated, negative regulation transcription from RNA polymerase promoter, protein binding, ATP binding, protein kinase binding, and chromatin binding. KEGG pathway analysis indicated that the DEmRNAs were significantly associated with PI3K-Akt, p53 signaling pathway, Hepatitis B, and many other tumor-related pathways such as miRNAs in cancer, pathways in cancer, colorectal cancer, small cell lung cancer, and prostate cancer.
After constructing a PPI network, 10 hub genes including CCNB1, E2F1, RRM2, and PBK were identified. Previous studies have reported that overexpression of CCNB1, PBK and RRM2 predicted poor survival in HCC, which may be potential therapeutic targets.43–45 EZH2 can initiate molecular processes such as NF-κB activation, miRNA silencing, and tumor immune evasion, which are considered to be the basic characteristics of cancer,46 and therefore, different types of EZH2 inhibitors have been developed for cancer therapy.47 ATAD2, an emerging oncogene closely related to the tumorigenesis in multiple cancers, has been validated to be a promising drug target for treatment development.48 As the hub genes are closely associated with tumorigenesis, we are interested in whether they can be used as potential good diagnostic indicators of AFP-negative HCC. The 10 hub genes identified in the present study demonstrated superior performance in the diagnosis of AFP-negative HCC, indicating their potential as diagnostic biomarkers for AFP-negative HCC.
In the present study, we identified the dysregulation RNAs and constructed a ceRNA network which may lead to the tumorigenesis in the AFP-negative HCC. Despite the results provide some candidate biomarkers for AFP-negative HCC, there are inevitably some limitations. More experimental validations are needed for clinical application. And whether there is a correlation between the ceRNA and HCC clinical progress needs more exploration.
Conclusion
In conclusion, a ceRNA was constructed and 2 lncRNAs (LINC00261, LINC00482), 3 miRNAs (hsa-miR-93, hsa-miR-221, hsa-miR-222), and 2 mRNAs (EGR2, LPCAT1) were associated with the prognosis of AFP-negative patients. And ten hub genes including EZH2, CCNB1, E2F1, PBK, CHAF1A, ESR1, RRM2, CCNE1, MCM4, and ATAD2 showed good diagnostic power in AFP-negative patients. Our research identified potential diagnostic and prognostic biomarkers for AFP-negative HCC, which provided novel insights into the tumorigenesis mechanism of AFP-negative HCC.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (No. 81773940 and No. 81473388).
Disclosure
The authors declared no conflicts of interest for this work and no competing financial interests.
References
1. She S, Xiang Y, Yang M, et al. C-reactive protein is a biomarker of AFP-negative HBV-related hepatocellular carcinoma. Int J Oncol. 2015;47(2):543–554. doi:10.3892/ijo.2015.3042
2. Cui DJ, Wu Y, Wen DH. CD34, PCNA and CK19 expressions in AFP- hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2018;22(16):5200–5205.
3. Huang X, Sun L, Wen S, et al. RNA sequencing of plasma exosomes revealed novel functional long noncoding RNAs in hepatocellular carcinoma. Cancer Sci. 2020;111(9):3338–3349. doi:10.1111/cas.14516
4. Luo CL, Rong Y, Chen H, et al. A logistic regression model for noninvasive prediction of AFP-negative hepatocellular carcinoma. Technol Cancer Res Treat. 2019;18:1533033819846632. doi:10.1177/1533033819846632
5. Jing W, Peng R, Zhu M, et al. Differential expression and diagnostic significance of pre-albumin, fibrinogen combined with D-dimer in AFP-negative hepatocellular carcinoma. Pathol Oncol Res. 2020;26(3):1669–1676. doi:10.1007/s12253-019-00752-8
6. Li MS, Ma QL, Chen Q, et al. Alpha-fetoprotein triggers hepatoma cells escaping from immune surveillance through altering the expression of Fas/FasL and tumor necrosis factor related apoptosis-inducing ligand and its receptor of lymphocytes and liver cancer cells. World J Gastroenterol. 2005;11(17):2564–2569. doi:10.3748/wjg.v11.i17.2564
7. Wang X, Mao M, He Z, et al. Development and validation of a prognostic nomogram in AFP-negative hepatocellular carcinoma. Int J Biol Sci. 2019;15(1):221–228. doi:10.7150/ijbs.28720
8. Lu LH, Zhang YF, Wei W, Shi M, Guo RP. Preoperative carbohydrate antigen 19-9: its neglected role in alpha-fetoprotein-negative hepatocellular carcinoma patients. J Gastrointest Surg. 2017;21(12):2025–2032. doi:10.1007/s11605-017-3528-5
9. Jiang Y, Tie C, Wang Y, et al. Upregulation of serum sphingosine (d18:1)-1-P potentially contributes to distinguish HCC including AFP-negative HCC from cirrhosis. Front Oncol. 2020;10:1759. doi:10.3389/fonc.2020.01759
10. Zhang N, Chen Y, Shen Y, Lou S, Deng J. Comprehensive analysis the potential biomarkers for the high-risk of childhood acute myeloid leukemia based on a competing endogenous RNA network. Blood Cells Mol Dis. 2019;79:102352. doi:10.1016/j.bcmd.2019.102352
11. Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67(3):603–618. doi:10.1016/j.jhep.2017.04.009
12. Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi:10.1038/nature09819
13. Zhou RS, Zhang EX, Sun QF, et al. Integrated analysis of lncRNA-miRNA-mRNA ceRNA network in squamous cell carcinoma of tongue. BMC Cancer. 2019;19(1):779. doi:10.1186/s12885-019-5983-8
14. Ba Z, Gu L, Hao S, Wang X, Cheng Z, Nie G. Downregulation of lncRNA CASC2 facilitates osteosarcoma growth and invasion through miR-181a. Cell Prolif. 2018;51(1). doi:10.1111/cpr.12409
15. Long J, Bai Y, Yang X, et al. Construction and comprehensive analysis of a ceRNA network to reveal potential prognostic biomarkers for hepatocellular carcinoma. Cancer Cell Int. 2019;19:90. doi:10.1186/s12935-019-0817-y
16. Abbastabar M, Sarfi M, Golestani A, Khalili E. lncRNA involvement in hepatocellular carcinoma metastasis and prognosis. EXCLI J. 2018;17:900–913.
17. Bai Y, Long J, Liu Z, et al. Comprehensive analysis of a ceRNA network reveals potential prognostic cytoplasmic lncRNAs involved in HCC progression. J Cell Physiol. 2019;234(10):18837–18848. doi:10.1002/jcp.28522
18. Zhang Z, Qian W, Wang S, et al. Analysis of lncRNA-associated ceRNA network reveals potential lncRNA biomarkers in human colon adenocarcinoma. Cell Physiol Biochem. 2018;49(5):1778–1791. doi:10.1159/000493623
19. Yan Y, Lu Y, Mao K, et al. Identification and validation of a prognostic four-genes signature for hepatocellular carcinoma: integrated ceRNA network analysis. Hepatol Int. 2019;13(5):618–630. doi:10.1007/s12072-019-09962-3
20. Guo D, Li Y, Chen Y, et al. DANCR promotes HCC progression and regulates EMT by sponging miR-27a-3p via ROCK1/LIMK1/COFILIN1 pathway. Cell Prolif. 2019;52(4):e12628. doi:10.1111/cpr.12628
21. Fan J, Zhang J, Huang S, Li P. lncRNA OSER1-AS1 acts as a ceRNA to promote tumorigenesis in hepatocellular carcinoma by regulating miR-372-3p/Rab23 axis. Biochem Biophys Res Commun. 2020;521(1):196–203. doi:10.1016/j.bbrc.2019.10.105
22. Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28(15):2062–2063. doi:10.1093/bioinformatics/bts344
23. Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43(Database issue):D146–D152. doi:10.1093/nar/gku1104
24. Chou CH, Chang NW, Shrestha S, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44(D1):D239–D247. doi:10.1093/nar/gkv1258
25. Fromm B, Billipp T, Peck LE, et al. System for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome. Annu Rev Genet. 2015;49:213–242. doi:10.1146/annurev-genet-120213-092023
26. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
27. Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–D815. doi:10.1093/nar/gks1094
28. Luo P, Wu S, Yu Y, et al. Current status and perspective biomarkers in AFP negative HCC: towards screening for and diagnosing hepatocellular carcinoma at an earlier stage. Pathol Oncol Res. 2020;26(2):599–603. doi:10.1007/s12253-019-00585-5
29. Bai DS, Zhang C, Chen P, Jin SJ, Jiang GQ. The prognostic correlation of AFP level at diagnosis with pathological grade, progression, and survival of patients with hepatocellular carcinoma. Sci Rep. 2017;7(1):12870. doi:10.1038/s41598-017-12834-1
30. Wang X, Gao C, Feng F, et al. Construction and analysis of competing endogenous RNA networks for breast cancer based on TCGA dataset. Biomed Res Int. 2020;2020:4078596.
31. Mo M, Liu S, Ma X, et al. A liver-specific lncRNA, FAM99B, suppresses hepatocellular carcinoma progression through inhibition of cell proliferation, migration, and invasion. J Cancer Res Clin Oncol. 2019;145(8):2027–2038. doi:10.1007/s00432-019-02954-8
32. Chen YA, Cheng L, Zhang Y, Peng L, Yang HG. LncRNA RUSC1-AS1 promotes the proliferation of hepatocellular carcinoma cells through modulating NOTCH signaling. Neoplasma. 2020;67(6):1204–1213. doi:10.4149/neo_2020_191010N1024
33. Shi Y, Yang X, Xue X, et al. HANR promotes hepatocellular carcinoma progression via miR-214/EZH2/TGF-beta axis. Biochem Biophys Res Commun. 2018;506(1):189–193. doi:10.1016/j.bbrc.2018.10.038
34. Xu C, He T, Li Z, Liu H, Ding B. Regulation of HOXA11-AS/miR-214-3p/EZH2 axis on the growth, migration and invasion of glioma cells. Biomed Pharmacother. 2017;95:1504–1513. doi:10.1016/j.biopha.2017.08.097
35. Xia H, Ooi LL, Hui KM. MicroRNA-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer. Hepatology. 2013;58(2):629–641. doi:10.1002/hep.26369
36. Wei Q, Zhao L, Jiang L, et al. Prognostic relevance of miR-137 and its liver microenvironment regulatory target gene AFM in hepatocellular carcinoma. J Cell Physiol. 2019;234(7):11888–11899. doi:10.1002/jcp.27855
37. Sakabe T, Azumi J, Umekita Y, et al. Prognostic relevance of miR-137 in patients with hepatocellular carcinoma. Liver Int. 2017;37(2):271–279. doi:10.1111/liv.13213
38. Xu J, Liu C, Zhou L, et al. Distinctions between clinicopathological factors and prognosis of alpha-fetoprotein negative and positive hepatocelluar carcinoma patients. Asian Pac J Cancer Prev. 2012;13(2):559–562. doi:10.7314/APJCP.2012.13.2.559
39. Zhao T, Jia L, Li J, et al. Heterogeneities of site-specific N-glycosylation in HCC tumors with low and high AFP concentrations. Front Oncol. 2020;10:496. doi:10.3389/fonc.2020.00496
40. Chen Z, Xiang L, Huang Y, Fang Y, Li X, Yang D. [Expression of long noncoding RNA linc00261 in hepatocellular carcinoma and its association with postoperative outcomes]. Nan Fang Yi Ke Da Xue Xue Bao. 2018;38(10):1179–1186. Chinese.
41. Zhang HF, Li W, Han YD. LINC00261 suppresses cell proliferation, invasion and Notch signaling pathway in hepatocellular carcinoma. Cancer Biomark. 2018;21(3):575–582. doi:10.3233/CBM-170471
42. Morita Y, Sakaguchi T, Ikegami K, et al. Lysophosphatidylcholine acyltransferase 1 altered phospholipid composition and regulated hepatoma progression. J Hepatol. 2013;59(2):292–299. doi:10.1016/j.jhep.2013.02.030
43. Yang QX, Zhong S, He L, et al. PBK overexpression promotes metastasis of hepatocellular carcinoma via activating ETV4-uPAR signaling pathway. Cancer Lett. 2019;452:90–102. doi:10.1016/j.canlet.2019.03.028
44. Zhou Z, Li Y, Hao H, et al. Screening hub genes as prognostic biomarkers of hepatocellular carcinoma by bioinformatics analysIS. Cell Transplant. 2019;28(1_suppl):76S–86S. doi:10.1177/0963689719893950
45. Yue C, Ren Y, Ge H, et al. Comprehensive analysis of potential prognostic genes for the construction of a competing endogenous RNA regulatory network in hepatocellular carcinoma. Onco Targets Ther. 2019;12:561–576. doi:10.2147/OTT.S188913
46. Yamagishi M, Uchimaru K. Targeting EZH2 in cancer therapy. Curr Opin Oncol. 2017;29(5):375–381. doi:10.1097/CCO.0000000000000390
47. Duan R, Du W, Guo W. EZH2: a novel target for cancer treatment. J Hematol Oncol. 2020;13(1):104. doi:10.1186/s13045-020-00937-8
48. Hussain M, Zhou Y, Song Y, et al. ATAD2 in cancer: a pharmacologically challenging but tractable target. Expert Opin Ther Targets. 2018;22(1):85–96. doi:10.1080/14728222.2018.1406921
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.