Back to Journals » International Journal of General Medicine » Volume 16
Investigation of Diagnostic and Prognostic Value of CLEC4M of Non-Small Cell Lung Carcinoma Associated with Immune Microenvironment
Authors Liu H, Yu Z, Liu Y, Li M, Chen C, Zhu Z, Liu F, Tan L
Received 10 January 2023
Accepted for publication 30 March 2023
Published 15 April 2023 Volume 2023:16 Pages 1317—1332
DOI https://doi.org/10.2147/IJGM.S397695
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Huan Liu,1,2,* Zhiping Yu,3,* Yueguang Liu,4 Mingzhen Li,1,2 Cheng Chen,1,2 Zhiyu Zhu,4 Fang Liu,4 Liming Tan2,3
1Department of Precision Medicine Center, The Second People’s Hospital of Huaihua, Huaihua, People’s Republic of China; 2Key Laboratory of Cancer Prevention and Treatment of Huaihua, Huaihua, People’s Republic of China; 3School of Pharmacy, Xuzhou Medical University, Xuzhou, People’s Republic of China; 4Department of Clinicopathology Center, The Second People’s Hospital of Huaihua, Huaihua, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Liming Tan, The Second People’s Hospital of Huaihua, Lulin Road, Huaihua, 418000, People’s Republic of China, Email [email protected]
Purpose: C-type lectin domain family 4 member M (CLEC4M) has been found to be involved in the occurrence and development of cancer, but its role in NSCLC remains to be fully explored. Our work aims to evaluate the diagnostic and prognostic value of CLEC4M in NSCLC and to investigate the underlying mechanisms of CLEC4M in the immune microenvironment of NSCLC.
Methods: Integrating publicly accessible data and clinical tissue samples to verify the expression of CLEC4M in NSCLC. The diagnostic value of CLEC4M was determined by receiver operating characteristic (ROC) curve. Kaplan–Meier survival analysis, nomogram plot, univariate and multivariate Cox regression models were performed to evaluate the prognostic impact of CLEC4M on NSCLC patients. The correlation between CLEC4M and tumor immune infiltration was estimated using TIMER and UALCAN databases. Functional assessments including GO, KEGG pathway and GSEA analyses were implemented to illustrate the potential mechanisms of CLEC4M in NSCLC.
Results: CLEC4M was significantly downregulated in NSCLC tissue, as confirmed by immunohistochemistry of clinical tissues. The high AUC value of ROC curves demonstrated the diagnostic accuracy of CLEC4M in NSCLC. Additionally, low CLEC4M expression was associated with poor survival in NSCLC patients. Furthermore, CLEC4M was found to be significantly associated with tumor immune infiltration, and CLEC4M may be involved in immune activation and proliferation inhibition through the functional assessment, suggesting that CLEC4M may be a therapeutic target for NSCLC patients.
Conclusion: Our findings reveal CLEC4M is significantly downregulated in NSCLC tissues, and illustrate the diagnostic and prognostic value of CLEC4M in NSCLC, as well as its potential serve as an immune-related therapeutic target.
Keywords: CLEC4M, non-small cell lung carcinoma, NSCLC, prognosis, diagnosis, tumor immune microenvironment
Introduction
Lung cancer (LC) is the most commonly diagnosed cancer and accounts for over 18% of cancer-related deaths worldwide. In patients with advanced stage of LC, the 5-year overall survival rate is less than 20%. In approximately 80% of lung cancer cases, the histological type is non-small cell lung carcinoma (NSCLC), which is mainly categorized into lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC).1–3 Increasing studies have revealed the vital impacts of genomic aberrations on the proliferation and progression of tumor cells.4,5 Thus, identification of novel reliable molecular signature and therapeutic targets to enhance treatment effectiveness, especially in NSCLC, is urgently demanded.
Accumulating evidence suggested that the interaction between tumor cells and the tumor microenvironment, specifically the immune microenvironment, is considered to play a key role in tumor progression. Over the past decade, cancer treatment has undergone a revolution, shifting away from drugs that broadly target tumors, such as chemotherapy and radiotherapy, to the use of antibody-based immunotherapy to modulate immune responses against tumors. Immune checkpoint blockade (ICB) works by blocking the interaction of receptors and/or molecular ligands, which are involved in suppressing T cell activation or function.6–8 ICB therapy has shown significant clinical benefit in a small number of patients with durable responses, such as the inhibitor of programmed cell death receptor-1 (PD-1) and its ligand (PD-L1) have shown promising antitumor effects in various cancers, including NSCLC.9 Additionally, increasing evidence shows the prognosis and therapy effectiveness can be affected by tumor-infiltrating immune cells.10,11 Therefore, it will be particularly important to reveal the immunophenotype of tumor-immune interactions and explore novel immune-related therapeutic targets.
Lately, several studies have reported that lectins may play a significant role in tumor progression and metastasis. A series of selectins such as E-, P-, and L-selectin can promote lung metastasis by the formation of metastasis microenvironment, which is mediated by the interactions between tumor cells and blood components.12 Other researchers confirmed the low level of mannose-binding lectin (MBL) raised the risk of gastric cancer, and elevated MBL level was notably correlated with adverse survival for patients with lung cancer.13 In addition, hepatic sinusoidal endothelial cell lectin (LSECtin) promotes the occurrence of liver metastases in colon cancer.14 CLEC4M, a Ca2+-dependent C-type lectin, also known as DC-SIGNR, L-SIGN or CD209L, functions in cell adhesion and pathogen recognition. CLEC4M protein is comprised of four distinct domains: a C-terminal carbohydrate recognition domain, a flexible tandem-repeat neck domain of variable length, a transmembrane region and an N-terminal cytoplasmic domain involved in internalization. CLEC4M is closely related to neighboring gene DC-SIGN in terms of sequence and function, but emerging evidence has proposed their distinct roles in immunity and tumor progression. CLEC4M is highly expressed in hepatic sinusoidal endothelial cells, lymph node endothelial cells and placental capillary endothelial cells, but not in peripheral blood-derived dendritic cells. Additionally, CLEC4M is also expressed in cytokeratin-positive alveolar epithelium, as well as in a group of cells that co-express angiotensin-converting enzyme-2 (ACE2) but is cytokeratin-negative.15–17 Our previous work has demonstrated that a positive association between CLEC4M expression and cell sensitivity in cisplatin-based chemotherapy of NSCLC.18 Other studies have shown that serum CLEC4M levels are lower in hepatocellular carcinoma (HCC) compared with normal controls.19 As a receptor for a variety of viruses including SARS-CoV, HIV, HCV, influenza A and tuberculosis viruses, CLEC4M has also been identified as a potential target related to tumorigenesis. However, the biological functions of CLEC4M in NSCLC have not been thoroughly investigated. This study mainly focused on examining the diagnostic and prognostic value of CLEC4M in NSCLC, and evaluating the role of CLEC4M in the tumor immune microenvironment of NSCLC.
Methods and Materials
Acquisition of NSCLC Expression Profiles from Publicly Available Database
NSCLC transcriptional profiles and the corresponding clinical data were retrieved from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov) and Gene Expression Omnibus (GEO) database, which contains lung adenocarcinoma (LUAD) and lung squamous cell cancer (LUSC) patients. The expression distributions of CLEC4M at the tumor and adjacent tissue were determined.20,21 The GEPAI2 tool (http://gepia2.cancer-pku.cn/) was also employed to reveal the difference of CLEC4M between normal and NSCLC tissue.22 In addition, NSCLC methylation data was obtained from the UALCAN database to assess the impact of CLEC4M genetic modification (http://ualcan.path.uab.edu/).23
Validation of CLEC4M Expression Using Immunohistochemistry Detection
A total of 13 NSCLC paraffin-embedded tissues were enrolled for this analysis. Tissues were deparaffinized with a histologic clearing agent and a series of rehydration ethanol washes. Antigen retrieval was performed by boiling samples in 10 mm sodium citrate buffer, pH 6, with 0.05% Tween 20 in a microwave for 20 min. To block endogenous peroxidase, each slide was incubated with 3% hydrogen peroxide for 30 min and then 10% normal goat serum was used to block. Sections were incubated overnight at 4°C with a specific primary antibody of CLEC4M (ab169783; Abcam, Cambridge, UK). Slides were rinsed with PBST and incubated with secondary antibody for 1 h at room temperature. Staining was visualized with 3, 3-diaminobenzidine, and counterstained with hematoxylin.
Estimation of CLEC4M-Related Immune Infiltration for NSCLC Patients
Estimation of Stromal and Immune cells in Malignant Tumor tissues using Expression data (ESTIMATE) is a practical approach to calculate the proportion of immune and stromal cells in tumor samples.24 The ESTIMATE algorithm was implemented based on gene expression profile downloaded from TCGA using R-package “estimate”. Then, LUAD and LUSC samples were categorized into high score group and low score group based on the median value of the calculated immune score.
Tumor immune estimation resource (TIMER, https://cistrome.shinyapps.io/timer) is a web tool designed to systematically analyse the immune infiltration level of tumor samples.25 With the TIMER algorithm, the abundances of six immune infiltrates (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells) are estimated. In addition, specific gene expression levels are also displayed against tumor purity. The “SCNA” module allows the comparison of tumor infiltration levels among tumors with different somatic copy number alterations for a given gene, displaying the distributions of each immune subset at each copy number status in tumor. Moreover, the “Correlation” module shows the expression relations between a pair of genes with scatterplots, whose statistical significance is estimated using Spearman’s rho value. These modules mentioned above were used to explore the association between CLEC4M expression and immune markers.
Investigation of Enriched Functions Across CLEC4M
A “coexpression” strategy to detect the expression-related gene of CLEC4M using Pearson’s correlation, these genes co-expressed with CLEC4M were screened by Pearson coefficient >0.4 and p value <0.05.26 Then, these CLEC4M-coexpressed genes were involved in the functional assessments including Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses.27,28 GO terms uncover the functions of candidate signatures, including biological processes (BPs), molecular functions (MFs), and cellular components (CCs). KEGG analysis reflects the pathways in which genes are enriched. The p value of <0.05 was considered as statistically significant.
Gene set enrichment analysis (GSEA) is a software to determine the concordance and difference between two biological states using a predefined gene set.29 LUAD and LUSC datasets obtained from TCGA were analyzed by GSEA to estimate the enriched function that responds to the CLEC4M expression. According to the median CLEC4M expression value, samples were split into a high CLEC4M expression group and a low CLCEM4 expression group. GSEA was performed using R package “clusterProfiler”. The threshold of statistical significance was set as p value <0.05.
Statistical Analysis
All the statistical analyses were implemented with R software (version 4.0). The prognostic value of CLEC4M in NSCLC was predicted using Kaplan–Meier survival analysis, followed by the log-rank p-value. A best cutoff point of CLEC4M expression was applied to divide the NSCLC samples into high expression group and low expression group, which was calculated by R package “survival”.30 Nomogram was plotted using R package “rms”. Receiver operating characteristic (ROC) curves were used to assess the diagnostic value of CLEC4M in NSCLC. The comparison of CLEC4M expression between different groups was explored using Wilcoxon test. The correlation between CLEC4M expression and clinical factors was analyzed by the Chi-square test. Univariate and multivariate analyses were carried out with a Cox proportional hazards model. Functional assessments of GO and KEGG pathway were analyzed using Fisher’s exact test.31 The p-value cutoff <0.05 was considered statistically significant.
Results
Down-Regulated Expression of CLEC4M in NSCLC
To determine the expression of CLEC4M in NSCLC, accessible public data from TCGA, GEO and GEPIA2 databases were used, in which the tumor and adjacent normal tissue of LUAD and LUSC were examined. The results indicated that CLEC4M was significantly downregulated in NSCLC, including LUAD and LUSC (Figure 1a). In addition, the CLEC4M expression profile of GEPIA2 database showed significantly lower level in both LUAD and LUSC than normal tissue (Figure 1b). Consistently, the expression data of CLEC4M from matched tumor and noncancerous tissue of LUAD (Figure 1c) and LUSC (Figure 1d) showed similar results to the above conclusions. The immunohistochemical staining of 13 clinical lung tumor patients confirmed the low expression of CLEC4M in tumor tissue, including LUAD and LUSC cases (Figure 1e–g). Furthermore, correlation analysis between CLEC4M expression and clinical characteristics was also performed. CLEC4M expression was found to associate with sex (p = 0.001), pathological stage (p = 0.029), T stage (p = 0.007) in TCGA (Figure 1h), and also with age (p = 0.044), N stage (0.016) and T stage (p = 0.034) in GSE30219 (Figure 1i).
CLEC4M Expression is Correlated with NSCLC Diagnosis and Prognosis
The diagnostic ability of CLEC4M in NSCLC was evaluated using TCGA and GEO datasets. As depicted, the results of ROC revealed the high diagnostic accuracy of CLEC4M in LUAD and LUSC cases of TCGA with AUC = 0.901, AUC = 0.918, respectively (Figure 2a and b). In addition, CLEC4M also showed significant diagnostic potential in LUAD and LUSC of GSE30219 with AUC = 0.715 and 0.703, respectively (Figure 2c and d). Moreover, the patients with early phase NSCLC (stage T1) were collected to evaluate the diagnostic value of CLEC4M. CLEC4M was found to be significantly reduced in stage T1 NSCLC tissues of GSE30219 (Figure 2f), and the result of ROC indicated significant diagnostic ability with AUC = 0.805 (Figure 2e). The above results demonstrate the robustness and accuracy of CLEC4M in the diagnosis of NSCLC patients.
To detect the prognostic value of CLEC4M in NSCLC, GEO datasets and Kaplan–Meier Plotter database were accessed to perform survival analysis, using Kaplan–Meier curve and Log rank test. Samples were grouped based on the best cutoff value of CLEC4M expression. The results illustrated that the decreased expression of CLEC4M was remarkably associated with poor OS of NSCLC patients in GSE30219 (p = 0.031) (Figure 3a). Similarly, the low CLEC4M expression was linked to worse OS of patients with LUAD and LUSC than those with high CLEC4M expression (LUAD, p = 0.042; LUSC, p = 0.012) (Figure 3b and c). In dataset GSE31210 and CAarray, the down-regulated expression of CLEC4M was correlated with adverse prognosis of NSCLC patients regarding OS (CAarray, p = 0.019; GSE31210, p = 0.0041) (Figure 3d and f), and progression-free survival (PFS: CAarray, p = 0.0053; GSE31210, p = 0.022) (Figure 3e and g). In addition, a nomogram was constructed according to the clinical features of GSE30219, suggesting the low CLEC4M expression may play as a risk factor to predict the prognosis of NSCLC patients (Figure 3h), and the dynamic nomogram was also deployed for such an exhibition (https://nomforclec4m.shinyapps.io/DynamicNomogram/).
To better explore the prognostic effect of CLEC4M expression on NSCLC survival, the association between CLCE4M expression and clinicopathological features was analyzed using the Kaplan–Meier plotter database. The decreased CLEC4M expression was tightly correlated with poor prediction of OS in stage 1, stage T1 and stage N0 to N1 patients. Additionally, the worse OS and PFS of patients with grades 1 to 2 could be predicted by the downregulation of CLEC4M expression (Table 1). Furthermore, univariate and multivariate COX regressive models were also constructed to estimate the prognostic potential of CLEC4M using datasets GSE30219. When compared with clinicopathological factors age, gender, stage T and N, CLEC4M showed a significantly prognostic potential in NSCLC (univariate analysis for LUAD, HR = 0.51, 95% CI 0.27–0.99, Cox p = 0.047. For LUSC, HR = 0.25, 95% CI 0.07–0.80, Cox p = 0.018; Multivariate analysis for LUAD: HR = 0.51, 95% CI 0.25–0.99, Cox p = 0.048. For LUSC, HR = 0.023, 95% CI 0.07–0.75, Cox p = 0.013) (Table 2 and Table 3), which further proposed that CLEC4M was a predictive indicator of survival in NSCLC. Thus, the lower CLEC4M expression can be identified as an independent risk factor to predict adverse outcome for NSCLC patients.
 |
Table 1 The Impact of CLEC4M on NSCLC Prognosis with Clinicopathological Factors in the Kaplan-Meier Plotter Database |
 |
Table 2 Univariate and Multivariate Cox Regression Analyses of LUAD Patients in GSE30219 Regarding Overall Survival |
 |
Table 3 Univariate and Multivariate Cox Regression Analyses of LUSC Patients in GSE30219 Regarding Overall Survival |
CLEC4M Expression is Related to Immune Infiltration in NSCLC Microenvironment
NSCLC patients with the same histological type may suffer different clinical outcomes, mainly due to differences in tumor immune infiltration. A higher level of lymphocytic tumor infiltration was indicated to result in a favorable prognosis for NSCLC. However, the mechanism remains completely uncovered. We assessed the association of CLEC4M expression between immune infiltration levels to reveal potential mechanisms for how CLEC4M affects NSCLC prognosis.
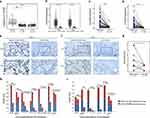
The ESTIMATE algorithm was deployed to calculate the immune score of NSCLC patients from TCGA cohort to evaluate the immune-related cell infiltration level in tumor, including LUAD and LUSC. Patients were divided into high- and low-score groups based on the median value of immune score, then, on which the distribution of CLEC4M expression was carried out. Apparently, patients with LUAD displayed elevated CLEC4M expression in the high immune score group, as was the case in LUSC, implying a strong correlation of CLEC4M expression with immune filtration in NSCLC patients (Figure 4a and b). Furthermore, TIMER was utilized to determine the association of CLEC4M expression with tumor purity and immune infiltration levels in NSCLC. As shown (Figure 4c and j), the expression of CLEC4M was negatively correlated to tumor purity in LUAD and LUSC, and positively correlated with the infiltration level of CD8+ T cells (Figure 4e and l), macrophages (Figure 4g and n), neutrophils (Figure 4h and o) and dendritic cells (Figure 4i and p). Disparately, the CLEC4M expression was positively associated with the infiltration of CD4+ T cells in LUSC (r = 0.205, p = 6.78e-06) (Figure 4m), but not in LUAD (r = 0.084, p = 0.066) (Figure 4f). As for B cells, the infiltration level was not correlated with the CELC4M expression (Figure 4d and k). These findings implicate a potential role of CLEC4M in immune infiltration of NSCLC.
A more detailed list was executed to analyze the association between CLEC4M and immune infiltration in LUAD and LUSC by calculating the correlation coefficients of CLEC4M with immune-related markers (Table 4). Generally, all of these immune gene markers were positively coexpressed with CLEC4M. In LUAD, CLEC4M was positively correlated with the gene expression of most functional T cells, including CD8+T cells, T helper cell 1 (Th1), helper T cell 2 (Th2), regulatory T cells and depletion T cells. Meanwhile, the expression of CLEC4M was significantly correlated to the marker genes of immune cells such as neutrophil, natural killer cells, tumor-associated macrophages (TAM), and M1/M2 macrophages. These results also affirmed the results in Figure 4 even after correcting the tumor purity. In addition, similar results were found in LUSC. Moreover, how the level of immune infiltration varied with the gene copy numbers of CLEC4M was analyzed using the SCNA module of TIMER. The results showed that alterations in the CLEC4M gene copy number were significantly associated with infiltration levels of immune cells, including B cells, CD4+T cells, macrophages, neutrophils, and DCs (Figure 4q), which provides a comprehensive view of the impact of CLEC4M on the immune system in NSCLC.
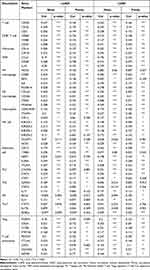 |
Table 4 The Correlations Between CLEC4M with Immune-Related Markers |
CLEC4M Plays a Role in Immune Activation and Proliferation Suppression in NSCLC
Functional enrichment analysis of GO term and KEGG pathway included a total of 209 genes coexpressed with CLEC4M in NSCLC cases of TCGA. The GO results uncovered these potential CLEC4M-interacted genes in BPs, including cell surface receptor signaling pathway, immune system process, and neutrophil chemotaxis. With regard to CCs, these co-expressed genes were predominantly in terms relevant to the plasma membrane. In the terms of MFs, the most enriched terms of CLEC4M co-expressed genes were carbohydrate binding, receptor activity and transmembrane signaling receptor activity (Figure 5a). Notably, some carcinogenesis pathways were found, such as PI3K-Akt signaling pathway, Ras signaling pathway and chemokine signaling pathway (Figure 5b). The above results suggest that CLEC4M may play a role in tumorigenesis and progression of NSCLC.
To further explore the impact of differential expression of CLEC4M on biological functions, GSEA analysis was conducted based on the strategy that the included genes without association limitation with CLEC4M, which offered a systematic and objective insight into the potential mechanism of CLEC4M in NSCLC. We analyzed LUAD and LUSC cases of TCGA with all the gene set files in ClusterProfiler, p value <0.05 was considered statistically significant. The results revealed several enrichment terms were related to cancer, among which few immune activation terms were found enriched in the high CLEC4M expression group of LUAD and LUSC, implying CLEC4M may inhibit the NSCLC progression through immune activation (Figure 5c and d), which was corresponded to previous results (Figure 4). In addition, some gene sets of proliferation regulation were enriched in low CLEC4M expression group of LUAD and LUSC, suggesting that CLEC4M may be involved in the process of proliferation inhibition in NSCLC (Figure 5e and f).
Discussion
The mortality rate of NSCLC patients worldwide remains high, and nearly half of lung cancer patients are not diagnosed until an advanced stage. Currently, owing to the unclear mechanisms related to the progression of malignant tumor, the therapeutic strategies for advanced patients are still lacking, which demands further exploration to discover novel therapeutic targets.
The tetrameric transmembrane protein receptors encoded by CLEC4s family are considered to be a vital class of molecules involved in immune response and cell adsorption. It has previously been reported that the expression levels of CLEC4s are different in several tumors.32,33 However, the expression and functions of CLEC4M in lung cancer have not been elucidated. In this study, the public data analysis and clinical sample validation revealed that CLEC4M was remarkably downregulated in NSCLC. In addition, CLEC4M was found to be significantly correlated with prognosis, a decrease in CLEC4M could predict poor OS and PFS in NSCLC. The univariate and multivariate cox regression analysis further confirmed the prognostic value of CLEC4M, suggesting that the low expression of CLEC4M could be considered as an independent risk indicator for NSCLC patients. The effect of CLEC4M on clinicopathological factors indicated the low expression level of CLEC4M was associated with worse clinical stage on T and N, which may support an intrinsically suppressive role of CLEC4M in NSCLC, and according to recent studies, CLEC4M acts as a tumor suppressor in hepatocellular cancer.19 Notably, high AUC values for the diagnosis of early stage NSCLC patients were obtained from ROC analysis, demonstrating the reliability of CLEC4M to distinguish NSCLC patients from healthy individuals. Therefore, these findings indicate that CLEC4M could serve as a potential biomarker for the diagnosis and prognosis of NSCLC.
In recent years, with the deepening of the understanding of tumor microenvironment, the relationship between immunity and tumor development has been continuously revealed, and the discovery of novel prognostic and diagnostic biomarkers has attracted much attention.34 Currently, immunotherapy is effectively used in clinical practice for several cancers, but there is still plenty of room for improvement. Studies have reported that immune abnormalities are closely related to the occurrence and development of tumors.35,36 Given the function of CLEC4M in immunity, its role in the immune microenvironment of NSCLC has great exploration value. In our study, CLEC4M was significantly associated with immune infiltration of CD8+T cells, neutrophils, macrophages and dendritic cells in NSCLC. Among these immune cells, T cell infiltration is usually related to a better clinical outcome.37 Dendritic cells are generally used as specific antigen presenting cells to exert immune effects.38 Neutrophils play an important role in the connection between inflammation and tumors, which are considered as one of therapeutic target in a variety of tumors.39 Previous studies suggest macrophages play a dual role in the tumor microenvironment, M1-type macrophages can promote anti-tumor effect through inflammation, while M2-type macrophages, also known as tumor associated macrophages (TAMs), through inhibiting inflammation to suppress tumor immune functions has recently attracted extensive attention.40–42
In this study, the copy number alteration of CLEC4M showed a marked impact on immune infiltration. Of note, the promotor methylation analysis displayed an obviously higher methylation level of CLEC4M in LUAD than that in normal tissue (Supplementary Figure 1a), but not in LUSC patients (Supplementary Figure 1b). These genetic modifications of CLEC4M may have important implications for the occurrence and development of NSCLC, laying a foundation for elucidating its mechanism. Furthermore, five CLEC4M-related regulators including U2AF2, HNRNPA1, SRSF1, EWSR1, and FUS were obtained through the ENCORI database (https://starbase.sysu.edu.cn).43 Among them, U2 sRNA auxiliary factor 2 (U2AF2), is a non-snRNA protein required for U2 snRNA binding to pre-mRNA branch sites. Studies have shown that U2AF2 is involved in Warburg effect and tumorigenesis via the AKT/mTOR signaling pathway in lung cancer.44 HNRNPA1 encodes a member of a ubiquitously expressed heterogeneous family of nuclear ribonucleoproteins implicated in mRNA metabolism and transport. HNRNPA1 has been reported to play a role in the progress and metastasis of colon cancer and bladder cancer.45 SRSF1 is a serine/arginine (SR)-rich family of pre-mRNA splicing factors, which exerts oncogenic roles in breast cancer and inhibits autophagy in lung cancer through BCL-x splicing and interaction.46 EWSR1 encodes a protein involved in gene expression, cell adhesion, and RNA processing, and EWSR1-related diseases include Ewing sarcoma and desmoplastic small round cell tumor.47 FUS is known as an oncogene, encoding a multifunctional protein component of hnRNP complex, and recently, a FUS-induced circular RNA ZNF609 was reported to promote tumorigenesis and progression in lung cancer.48 These findings may underline the potential interactions between CLEC4M and above regulators, which are pivotal for the development of NSCLC and require further in vivo/vitro validation.
Through the enriched function analysis, we found that CLEC4M may play an important role in the activation and response of immune cells, comprising T cells, lymphocytes, myeloid leucocytes, macrophages, further confirming the crucial role of CLEC4M in immune activity, and uncovered the potential mechanism by which CLEC4M affected NSCLC microenvironment. Recently, Ye et al49 integrated CNV-mediated transcriptional networks, mRNA co-expression, and protein co-expression networks to reveal associations between multi-omics network centrality and NSCLC oncogenesis, proliferation, and patient survival, demonstrating the immunotherapy targets including PD1, PDL1, CTLA4 and CD27, were the top hub genes in most of the constructed multi-omics networks in NSCLC tumors. In Yu’s study,50 SPP1 was identified as a biomarker of NSCLC, which has significant coexpression relation to PD1 and PDL1, and acted as an immune-related target. In addition, Qiu et al51 proposed a novel apaQTL mechanism to investigate how apaQTL-SNPs in non-coding regions affect NSCLC risk, and apaQTL-SNP rs10138506 in CHURC1 was shown to correlate with longer mRNA 3’UTR length to promote tumor development. These novel findings can provide new insight into the association between genetic factors and the tumor microenvironment, and broaden our thoughts to further investigate the role of CLEC4M in the development and progression of NSCLC. We detected the correlation between CLEC4M and immune checkpoint markers to evaluate the potential of CLEC4M in anti-tumor effect. These classic immune checkpoint genes including PDCD1, CD274, CTLA4 and PDCD1LG2, of which the expression was positively associated with CLEC4M in LUAD and LUSC, implying a certain value of CLEC4M to against tumor development (Supplementary Figure 2a–h). Taken together, CLEC4M may serve as a therapeutic target for NSCLC patients.
Conclusion
In summary, CLEC4M was significantly downregulated in NSCLC and was remarkably associated with poor prognosis. In addition, CLEC4M can be used as a diagnostic marker for early NSCLC patients. Furthermore, CLEC4M was involved in immune activation and proliferation suppression, implying CLEC4M plays a key role in the development of NSCLC. What is more, CLEC4M exhibits the potential of anti-tumor with close link to immune checkpoints. All of these findings suggest that CLEC4M may be identified as a possible biomarker for diagnosis and prognosis of patient with NSCLC, as well as an immune-related therapeutic target. Our work provided a novel target and direction for NSCLC treatment. However, further investigations are required to confirm these results and explain the underlying mechanism of CLEC4M in NSCLC.
Data Sharing Statement
The datasets used in this study are available in publicly online repositories, the accession numbers have been mentioned in this article.
Ethics Approval and Consent to Participate
This study was approved by the Ethics Committee of The Second People’s Hospital of Huaihua and was conducted in accordance with the Declaration of Helsinki. All of the patients signed informed consent form before they participated in the study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas, took part in drafting, revising or critically reviewing the article, gave final approval of the version to be published, have agreed on the journal to which the article has been submitted, and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Scientific Research Fund of Hunan Provincial Health Commission (grant No. 202104081730), Science and Technology Program Fund of Huaihua (grant No. 2021R3111) and the Fund of Huaihua Technology Innovation platform (grant No. 2021R2206).
Disclosure
The authors declare no potential conflicts of interest.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi:10.3322/caac.21660
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:182.
3. Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi:10.1056/NEJMoa040938
4. Allegretti M, Fabi A, Buglioni S, et al. Tearing down the walls: FDA approves next generation sequencing (NGS) assays for actionable cancer genomic aberrations. J Exp Clin Cancer Res. 2018;37. doi:10.1186/s13046-018-0702-x
5. Zhou N, Sepesi B, Leung CH, et al. Impact of genomic aberrations and additional therapies on survival outcomes of patients with operable non-small cell lung cancer (NSCLC) from the NEOSTAR study. J Clin Oncol. 2021;39:8542. doi:10.1200/JCO.2021.39.15_suppl.8542
6. Postow MA, Sidlow R, Hellmann MD. Immune‐related adverse events associated with immune checkpoint blockade. N Engl J Med. 2018;378:158–168. doi:10.1056/NEJMra1703481
7. Kubli SP, Berger T, Araujo DV, Siu LL, Mak TW. Beyond immune checkpoint blockade: emerging immunological strategies. Nat Rev Drug Discov. 2021;20:899–919. doi:10.1038/s41573-021-00155-y
8. Morad G, Helmink BA, Sharma P, Wargo JA. Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell. 2021;184:5309–5337. doi:10.1016/j.cell.2021.09.020
9. Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science. 2015;348:124–128. doi:10.1126/science.aaa1348
10. Kim AR, Choi KS, Kim M-S, et al. Absolute quantification of tumor-infiltrating immune cells in high-grade glioma identifies prognostic and radiomics values. Cancer Immunol Immunother. 2021;70:1995–2008. doi:10.1007/s00262-020-02836-w
11. Zhang Y, Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol. 2020;17:807–821. doi:10.1038/s41423-020-0488-6
12. Ivetic A, Hoskins Green HL, Hart SJ. L-selectin: a major regulator of leukocyte adhesion, migration and signaling. Front Immunol. 2019;10. doi:10.3389/fimmu.2019.01068
13. Pine SR, Mechanic LE, Ambs S, et al. Lung cancer survival and functional polymorphisms in MBL2, an innate-immunity gene. J Natl Cancer Inst. 2007;99(18):1401–1409. doi:10.1093/jnci/djm128
14. Zhang Y, Feng Z, Xu Y, et al. Novel roles of LSECtin in gastric cancer cell adhesion, migration, invasion, and lymphatic metastasis. Cell Death Dis. 2022;13:157.
15. Swystun LL, Notley CRP, Georgescu I, James PD, Lillicrap D. The endothelial lectin receptor CLEC4M internalizes factor VIII and von Willebrand factor via a clathrin-coated pit-dependent mechanism. Blood. 2013;122:1091. doi:10.1182/blood.V122.21.1091.1091
16. Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi:10.1126/science.abb2762
17. Ishibashi M, Morita N, Nomura-Kawaguchi C, Shimizu YK, Wakita T, Esumi M. CLEC4M-positive and CD81-negative Huh7 cells are not susceptible to JFH-1 HCVcc infection but mediate transinfection. Arch Virol. 2014;159:2949–2955. doi:10.1007/s00705-014-2150-z
18. Tan L-M, Li X, Qiu C-F, et al. CLEC4M is associated with poor prognosis and promotes cisplatin resistance in NSCLC patients. J Cancer. 2019;10:6374–6383. doi:10.7150/jca.30139
19. Yu Q, Gao K. CLEC4M overexpression inhibits progression and is associated with a favorable prognosis in hepatocellular carcinoma. Mol Med Rep. 2020;22:2245–2252. doi:10.3892/mmr.2020.11336
20. Colaprico A, Silva TC, Olsen C, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71–e71. doi:10.1093/nar/gkv1507
21. Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41:D991–D995 . doi:10.1093/nar/gks1193
22. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560 . doi:10.1093/nar/gkz430
23. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi:10.1016/j.neo.2017.05.002
24. Deng X, Lin -D-D, Zhang X, et al. Profiles of immune‐related genes and immune cell infiltration in the tumor microenvironment of diffuse lower‐grade gliomas. J Cell Physiol. 2020;235:7321–7331. doi:10.1002/jcp.29633
25. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi:10.1158/0008-5472.CAN-17-0307
26. Nahler G. Pearson correlation coefficient. Definitions. 2020;2020:1–4.
27. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi:10.1038/75556
28. Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114 . doi:10.1093/nar/gkr988
29. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi:10.1073/pnas.0506580102
30. Groeneveld CS, Chagas VS, Jones SJM, et al. RTNsurvival: an R/Bioconductor package for regulatory network survival analysis. Bioinformatics. 2019;35:4488–4489. doi:10.1093/bioinformatics/btz229
31. Routledge RD. Fisher’s exact test. Definitions. 2020;2020:524–525.
32. Zhang Y, Wei H, Fan L, et al. CLEC4s as potential therapeutic targets in hepatocellular carcinoma microenvironment. Front Cell. 2021;9:54.
33. Pees B, Yang W, Kloock A, et al. Effector and regulator: diverse functions of C. elegans C-type lectin-like domain proteins. PLoS Pathog. 2021;17:e1009454. doi:10.1371/journal.ppat.1009454
34. Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–550. doi:10.1038/s41591-018-0014-x
35. Baghban R, Roshangar L, Jahanban-Esfahlan R, et al. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun Signal. 2020;18:147.
36. Gajewski TF, Schreiber H, Fu Y-X. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014–1022. doi:10.1038/ni.2703
37. Liu Y, Sun Z. Turning cold tumors into hot tumors by improving T-cell infiltration. Theranostics. 2021;11:5365–5386. doi:10.7150/thno.58390
38. Banchereau JF, Steinman RM. Dendritic cells and the control of immunity. Nat Genet. 1998;392:245–252.
39. Németh T, Sperandio M, Mócsai A. Neutrophils as emerging therapeutic targets. Nat Rev Drug Discov. 2020;19:253–275. doi:10.1038/s41573-019-0054-z
40. Martínez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. Prime Rep. 2014;6:24.
41. Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–964. doi:10.1038/nri1733
42. Shapouri-Moghaddam A, Mohammadian S, Vazini H, et al. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol. 2018;233:6425–6440. doi:10.1002/jcp.26429
43. Li J, Liu S, Zhou H, Qu L, Yang J. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97 . doi:10.1093/nar/gkt1248
44. Laliotis GI, Chavdoula ED, Paraskevopoulou MD, et al. RETRACTED ARTICLE: AKT3-mediated IWS1 phosphorylation promotes the proliferation of EGFR-mutant lung adenocarcinomas through cell cycle-regulated U2AF2 RNA splicing. Nat Commun. 2021;12. doi:10.1038/s41467-021-24795-1
45. Chen C, Lin T, Huang J. MP01-06 exosome-transmitted long non-coding RNA PLAT1 recruits HNRNPA1 to promote lymphatic metastasis of bladder cancer. J Urol. 2020;205:1415–1420. doi:10.1097/JU.0000000000001545
46. Lv Y, Zhang W, Zhao J, et al. SRSF1 inhibits autophagy through regulating Bcl-x splicing and interacting with PIK3C3 in lung cancer. Signal Transduct. 2021;6:36.
47. Flucke UE, van Noesel MM, Siozopoulou V, et al. EWSR1—the most common rearranged gene in soft tissue lesions, which also occurs in different bone lesions: an updated review. Diagnostics. 2021;11:1093. doi:10.3390/diagnostics11061093
48. Liu S, Yang N, Jiang X, Wang J, Dong J, Gao Y. FUS‐induced circular RNA ZNF609 promotes tumorigenesis and progression via sponging miR‐142‐3p in lung cancer. J Cell Physiol. 2021;236:79–92. doi:10.1002/jcp.29481
49. Ye Q, Guo NL. Hub genes in non-small cell lung cancer regulatory networks. Biomolecules. 2022;12(12):1782. doi:10.3390/biom12121782
50. Yu L, Liang X, Wang J, et al. Identification of key biomarkers and candidate molecules in non-small-cell lung cancer by integrated bioinformatics analysis. Genet Res (Camb). 2023;2023:6782732. doi:10.1155/2023/6782732
51. Qiu A, Xu H, Mao L, et al. A novel apaQTL-SNP for the modification of non-small-cell lung cancer susceptibility across histological subtypes. Cancers. 2022;14(21):5309. doi:10.3390/cancers14215309
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.