Back to Journals » International Journal of Nanomedicine » Volume 11
Intracellular redox-responsive nanocarrier for plasmid delivery: in vitro characterization and in vivo studies in mice
Authors Zhang L, Zhang Y, Chen Z, He Y
Received 23 August 2015
Accepted for publication 7 January 2016
Published 11 October 2016 Volume 2016:11 Pages 5245—5256
DOI https://doi.org/10.2147/IJN.S94995
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Lei Yang
Lifen Zhang,1,2 Yushun Zhang,1,2 Zhenzhen Chen,3 Yuling He1
1State Key Laboratory of Applied Organic Chemistry, 2Key Laboratory of Nonferrous Metals Chemistry and Resources Utilization of Gansu Province, College of Chemistry and Chemical Engineering, Lanzhou University, Lanzhou, 3Department of Bioengineering, Zhengzhou University, Zhengzhou, Henan, People’s Republic of China
Abstract: Although some modifications of polyethyleneimine (PEI) properties have been explored to balance the transfection efficiency and cytotoxicity, its successful plasmid delivery in vitro and in vivo to realize its true therapeutic potentials remains a major challenge, mainly due to intracellular trafficking barriers. Herein, we present a delivery nanocarrier Pluronic-PEI-SS by conjugating reducible disulfide-linked PEI (PEI-SS) to biocompatible Pluronic for enhanced DNA delivery and transfection efficiency in vitro and in vivo. Pluronic-PEI-SS strongly condensed plasmid DNA to low positively charged nanocomplexes, exhibited good stability against deoxyribonuclease I digestion, and tended to be easily degraded in the presence of reducing agent 1,4-dithiothreitol. The in vitro transfection of the complex Pluronic-PEI-SS/DNA into HeLa and 293T cells resulted in lower cytotoxicity as well as significantly higher cellular uptake, nucleus transfection, and gene expression than Pluronic-PEI (25 kDa), PEI-SS, and PEI 25 kDa given alone. Furthermore, the in vivo transfection study demonstrated that Pluronic-PEI-SS/DNA complexes induced a higher enrichment than the commercial PEI/DNA complex in the tumor, indicating their potential application as biocompatible vector in gene delivery.
Keywords: responsive, gene delivery, polycation, Pluronic, disulfide-linked
Introduction
Gene therapy has gained significant attention over the past 2 decades as a promising approach for treating genetic disorders such as severe combined immunodeficiency1 and cystic fibrosis,2 as well as an alternative method to traditional chemotherapy used in treating cancer.2–4 Nucleic acids are easily degraded by enzymes within a couple of hours due to their susceptibilities to nuclease attacks.5 Therefore, it is necessary to use appropriate delivery carriers for DNA to enable enhanced translocation into cells and protection against enzyme-mediated degradation. Cationic polymer, polyethyleneimine (PEI), as a superior nonviral gene vector,6 was found to effectively condense plasmid, protect gene from degradation, transfect DNA into cells, and rupture the endosome because of its “proton sponge” nature.7 Unfortunately, there are still severe drawbacks for PEI/DNA polyplexes, such as serious cytotoxicity and poor DNA release, which reduce their transfection efficiencies and, especially, limit their potential applications in in vivo cancer therapy.8,9
In order to balance the transfection efficiency and cytotoxicity, some modifications of PEI properties were explored. Up to now, PEI has been modified with chloroquine, polyethylene glycol, heparin, folic acid, Pluronic (poly[ethylene oxide]-b-poly[propylene oxide]-b-poly[ethylene oxide]), and so on.10 For example, hydrophilic Pluronic polymers can possess the unique ability to incorporate into cell membranes as a result of the presence of the hydrophobic poly(propylene oxide) chain, as well as particular efficiency for transferring DNA in vivo, and good biocompatibility.11–18 It was reported that the overall gene transfection efficiency of polyplexes formulated with Pluronic-grafted PEI (25 kDa) was far higher than those formulated with Pluronic-grafted PEI (2 kDa) and unmodified PEI (25 kDa). However, their inefficient DNA release caused their low in vitro and in vivo gene transfection efficiency due to the strong binding between PEI 25 kDa segments and DNA.19
Therefore, in this study, novel bioreducible copolymer consisting of Pluronic and disulfide-containing low-molecular-weight PEI-SS was prepared via a simple conjugation. PEI-SS was used to facilitate DNA release, which can be readily triggered by internal factors (eg, glutathione [GSH]),20 and tend to be reduced and cleaved to low-molecular-weight linear PEI chains. Then, DNA would be quickly released from the complexes due to the weak electronic interaction of PEI short chains with DNA due to the reducibility of the PEI-SS (Figure 1). We investigated the DNA binding affinities, stability, degradability dependancy on reducing agent 1,4-dithiothreitol (DTT), cytotoxicity, and especially detailed in vitro and in vivo transfection efficacy of the nanocarrier Pluronic-PEI-SS, compared to PEI-SS, Pluronic-PEI (25 kDa) copolymer, and commercial 25 kDa PEI. The results demonstrated that Pluronic-PEI-SS is capable of reducing triggered DNA release and exhibits highly improved efficiency in DNA delivery both in vitro and in vivo, indicating its potential application as biocompatible vector in gene delivery.
Materials and methods
Materials
Linear PEI (MW =600 Da), branched commercial PEI (MW =25 kDa), Pluronic P123, DTT, dimethyl sulfoxide (DMSO), Dulbecco’s phosphate-buffered saline (PBS), cystamine dihydrochloride, 1,1′-carbonyldiimidazole, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), and Dulbecco’s Modified Eagle’s Medium (DMEM) were all obtained from Sigma-Aldrich Co. (St Louis, MO, USA). Acryloyl chloride was obtained from Rionlon Bohua (Tianjin) Pharmaceutical and Chemical Co, Ltd. (Tianjin, People’s Republic of China). Penicillin-streptomycin and fetal bovine serum (FBS) solutions were supplied from Thermo Fisher Scientific (Waltham, MA, USA). Plasmid pBR322 DNA and pGL4-control reporter gene were purchased from Fermentas China Co., Ltd. (Shenzhen, Guangdong, People’s Republic of China). Plasmid enhanced green fluorescent protein (pEGFP) containing the early promoter of Cytomegalovirus and enhanced green fluorescent protein (EGFP) was kindly donated by Professor Liang (Jianghan University). pEGFP was conjugated with 2-([1E,3E,5E]-5-[1-{5-carboxypentyl}-3,3-dimethylindolin-2-ylidene]penta-1,3-dienyl)-1-ethyl-3,3-dimethyl-3H-indolium chloride (Cy5) according to the protocol of the Label IT® Tracker™ intracellular nucleic acid localization kit (Mirus Bio LLC, Madison, WI, USA). Male nude mice (4–5 weeks old) were purchased from the Shanghai Experimental Animal Center (People’s Republic of China). All animal experiments complied with the requirements of the National Act on the Use of Experimental Animals (People’s Republic of China). This study was approved by the Medical Ethics Committee of The First Hospital of Lanzhou University.
Synthesis of Pluronic-PEI-SS conjugate copolymer and Pluronic-PEI (25 kDa)
PEI-SS was synthesized first according to conventional Schotten–Baumann conditions.21 Briefly, equimolar acryloyl chloride and cystamine were dissolved in dichloromethane at room temperature using trimethylamine as a catalyst. After the reaction was completed, cystamine bisacrylamide was obtained by filtration, purified by crystallization, and dried under vacuum. Then, the disulfide PEI-SS was synthesized through Michael addition between cystamine bisacrylamide and 600 Da PEI. The structures of PEI-SS were confirmed by proton nuclear magnetic resonance (1H NMR) and gel permeation chromatography–multi-angle laser light scattering (GPC-MALLS).
Pluronic-PEI-SS and Pluronic-PEI were synthesized by grafting Pluronic P123 to the amino groups of PEI-SS and PEI 25 kDa, as described previously.22 Briefly, the dissolved P123 (1.25 g) in tetrahydrofuran (THF, 15 mL) was slowly added to a solution of anhydrous THF containing carbonyldiimidazole (0.81 g) in a dropwise manner. After the reaction was carried out for 6 hours at room temperature under nitrogen atmosphere, 1,1-carbonyldiimidazole-activated P123 was obtained by precipitating three times in ice-cold diethyl ether. Then, the activated P123 in 15 mL anhydrous THF was allowed to react with PEI-SS at room temperature under nitrogen atmosphere for 48 hours. The resulting Pluronic-PEI-SS conjugate copolymers were dialyzed using Spectra/Por@ Biotech Dialysis membrane (MWCO 14 kDa), lyophilized, and characterized by 1H NMR and GPC-MALLS. Pluronic-PEI (25 kDa) copolymer was synthesized by the conjugation of activated P123 with PEI (25 kDa) according to similar procedure.
Degradation of Pluronic-PEI-SS
Behavior of the copolymer Pluronic-PEI-SS degradation in the presence of DTT was investigated by GPC-MALLS, using Pluronic-PEI as control. In brief, each copolymer (1 g) dissolved in PBS (2 mL, pH 7.4) was incubated with or without DTT (310 mM) at 37°C with shaking at 100 rpm for specified times. Then, the samples were frozen immediately in liquid nitrogen, lyophilized, and measured by GPC-MALLS to determine their molecular weights.
Preparation of complexes
To form polymer (Pluronic-PEI-SS, Pluronic-PEI, or PEI 25 kDa)/DNA complexes, pDNA (1 μg) dissolved in PBS (pH 7.4) was mixed with various amounts of polymer in PBS at various N/P (nitrogen to phosphate) ratios, vortexed for a short time, and further incubated at room temperature for 30 minutes.
Gel retardation assay
DNA condensing ability of Pluronic-PEI-SS, Pluronic-PEI, or PEI 25 kDa was examined by agarose gel electrophoresis. The polymer/DNA complexes at various N/P ratios were prepared and dissolved separately in 0.8% agarose at pH 7.5 by vigorous pipetting. The mixtures were incubated for 30 minutes at room temperature. Then, the mixture was electrophoresed on 0.8% agarose gel stained with ethidium bromide (0.1 μg/mL) at 80 V for 80 minutes, visualized by an ultraviolet (UV) (254 nm) illuminator and photographed with a gel imaging system (Kodak In-Vivo Imaging System FX Pro; Kodak, Rochester, NY, USA).
The stability of Pluronic-PEI-SS/DNA complex against deoxyribonuclease I (DNase I) digestion and reducing agent DTT was further investigated, respectively, using DNA as control. The samples were divided into three groups. The first group was treated with DNase I (1–2 units) for 30 minutes at 37°C. The second group was treated with different amounts of reductive reagent DTT (0, 0.5, 2.5, 12.5, 62.5, and 310 mM). The treated and untreated samples were then loaded on 0.8% agarose gel, stained with ethidium bromide, and photographed by gel imaging system.
Particle size and zeta potential measurements
The particle sizes and surface charges of complexes at various N/P ratios were measured at room temperature using a Zetaplus Zeta Potential Analyzer (Brookhaven Instrument Corp., Holtsville, NY, USA) equipped with a 15 mV solid-state laser operated at a wavelength of 635 nm and an angle of 90°.
Transmission electron microscopy
The morphologies of the Pluronic-PEI-SS/DNA complexes (N/P of 20) dependent on DTT were assessed by transmission electron microscopy (TEM). A drop of the solution with (or without) 310 mM DTT was placed on a Formvar precoated carbon grid for 30 minutes. Then, the samples were stained with 1% phosphotungstic acid for 15 minutes and analyzed using a JEM-1230 TEM microscope (JEOL, Tokyo, Japan) at 100 kV.
MTT assay
Human embryonic kidney transformed 293 (293T) and human cervical carcinoma cell line (HeLa) were incubated in DMEM with 10% FBS and 1% antibiotic solution (penicillin-streptomycin, 10,000 U/mL) at 37°C in a humidified atmosphere containing 5% CO2. Herein, MTT assay was carried out to evaluate the cytotoxicity of the complexes for different HeLa and 293T cells. Briefly, the cells were seeded in a 96-well plate at a density of 7,000 cells/well, and then incubated in 100 mL of DMEM with 10% FBS under a humidified atmosphere of 95% air and 5% CO2 for 24 hours. After that, the medium was replaced with 90 μL of fresh medium and the solutions (10 μL) of 25 kDa PEI, PEI-SS, Pluronic-PEI, or Pluronic-PEI-SS were added. After 48-hour treatment, MTT reagent (5 mg/mL, 10 μL in PBS) was injected into each well for incubation for another 4 hours at 37°C. To each well was added 100 μL of DMSO to dissolve the crystals, and then the medium was removed and DMSO (100 mL) was added and the absorbance of the solution was measured at 570 nm via a microplate reader (model 550; Bio-Rad Laboratories Inc., Hercules, CA, USA). The relative cell viability was calculated as follows: Viability (%) = (ODsample − ODblank)/(ODcontrol − ODblank) ×100, where ODsample is the absorbance of the solution of the cells cultured with the polymer or PEI, ODblank is the absorbance of the medium, and ODcontrol is the absorbance of the solution of the cells cultured with the medium only.
In vitro transfection
For transfection experiment, pEGFP was used to assess the transfection efficiency of the complexes including Pluronic-PEI-SS, PEI-SS, Pluronic-PEI, and 25 kDa PEI. Two different cells of HeLa and 293T were seeded in a 24-well plate at a density of 5×104 cells/well. The cells were incubated at 37°C in a humidified atmosphere of 95% air and 5% CO2 for 24 hours. The medium in each well was replaced with DMEM (1 mL) without FBS. The complexes with the N/P ratios from 10 to 40 (the amount of DNA added to each well was kept at 1 μg) were added to the cells and incubated for 4 hours at 37°C. The medium was then replaced with fresh DMEM medium (1 mL) with 10% FBS, for further 48-hour incubation. After that, the cells were separately harvested from wells by treatment with trypsin-ethylenediaminetetraacetic acid and suspended in microcentrifuge tubes with 1 mL PBS. The cells expressing green fluorescent protein were enumerated by fluorescence-activated cell sorting (FACSCanto™; BD Biosciences, San Jose, CA, USA).
Subcellular distribution
HeLa cells were seeded into six-well plates, 1×105 cells every well, and then cultured overnight in a cell incubator. The indicated complexes were mixed with Cy5-labeled plasmid and incubated for 30 minutes. For transfection, cell medium was changed with fresh complete medium, and then the complexes were added into the above medium. This was followed by 4 hours’ incubation. To examine the subcellular distribution, confocal and real-time polymerase chain reaction (PCR) assays were performed. The cells were stained with 4′,6-diamidino-2-phenylindole (DAPI), followed by observation with confocal laser scanning microscopy (Olympus FV1000; Olympus, Tokyo, Japan). Other cells were collected and used for nucleocytoplasmic separation, and then real-time PCR was performed using nuclear and cytoplasmic fractions as template.
In vivo transfection and expression
Male nude mice 4–5 weeks old and weighing 18–22 g were separated into three groups (five mice per group), and then subcutaneously injected with 2.4×106 HeLa cells to establish the tumor models. When the size of the transplanted tumors increased to 10 mm in diameter, at the 20th day, 200 μL of the sterilized complexes solution containing 50 μg of the pGL4-control or EGFP reporter gene was injected into the mice through the tail veins. The details of the complexes injected were as follows: Group 1, Pluronic-PEI-SS/DNA complexes with the N/P of 20; Group 2, PEI 25 KDa/DNA complexes with the N/P of 10; Group 3, free DNA. Twenty-four hours postinjection, the mice were sacrificed to collect the tumors. The tissues such as heart, liver, spleen, kidney, brain, and tumors were collected and homogenized in a cell lysis buffer. The cell lysate was centrifuged. After being centrifuged for 10 minutes at 10,000 rpm, an aliquot of the supernatant with pGL4-control was assayed for luciferase activity using a commercial kit and photon counting was performed with a luminometer.
The above collected the tumors expressing GFP protein, and embedded them in Tissue-Tek OCT, cooled quickly to −80°C, and sectioned with cryostat. The sections with size of 20 mm were put on Superfrost microscope slides and dried for 1 hour at room temperature and then kept at −80°C for subsequent use. The GFP fluorescence (excitation 488 cm−1) of the obtained samples was assessed by Zeiss 410 confocal laser scanning microscope equipped with an argon-krypton laser.
Statistical analysis
The statistical calculations between two sets of data were carried out using Student’s t-test software and differences were considered significant if P≤0.05.
Results and discussion
Synthesis and characterization of Pluronic-PEI-SS conjugate copolymer
The synthesis route for Pluronic-PEI-SS is illustrated in Figure 1. PEI-SS was synthesized by conjugating PEI and cystamine bisacrylamide. Figure 2A shows the 1H NMR spectrum of PEI-SS to confirm its chemical structure. The characteristic proton peaks between 2.5 and 3.0 ppm may be attributed to the special shift of PEI. It was found that acrylamide peaks between 5.8 and 6.5 ppm disappeared, indicating that PEI-SS was successfully prepared. Pluronic-PEI-SS (or Pluronic-PEI) was purified by dialysis to remove the unreacted PEI-SS (or PEI), unconjugated Pluronic, and other small molecules. The structures of Pluronic-PEI and Pluronic-PEI-SS were also determined by the 1H NMR spectra (Figure 2B and C).
The calculated composition of Pluronic to PEI-SS is ~0.5. The molecular weights and polydispersity indices of the three polymers were determined by GPC-MALLS. As shown in Table 1, the characteristics of the synthesized polymers were obtained, and the number average molecular weights (Mn) of Pluronic-PEI-SS and Pluroic-PEI were ~27 and 26 kDa, respectively.
Degradation of Pluronic-PEI-SS
Generally, disulfide bonds are susceptible to cleavage in the presence of DTT at physiological conditions.19 Herein, the degradation of Pluronic-PEI-SS was determined by its molecular weight changes, dependent on DTT, at 37°C, using Pluronic-PEI as control. Figure 3 displays the molecular weight profiles of the copolymers with (or without) DTT as a function of time. It was found that in the absence of DTT, Pluronic-PEI-SS hardly degraded and had little difference with Pluronic-PEI, while in the presence of DTT, Pluronic-PEI-SS degraded to a greater extent than Pluronic-PEI. Pluronic-PEI-SS was almost completely degraded within 10 hours in the presence of DTT and the molecular weights were kept relatively constant at ~5,000 Da even at 50 hours. These results indicate that Pluronic-PEI-SS including disulfide bonds has a rapid degradation in the reductive conditions and would display a rapid cleavage in the cytosol, which facilitates DNA release from the complex in gene delivery system.
Characterizations of the complexes
The Pluronic-PEI-SS/DNA complexes at various N/P ratios ranging from 0 to 40 were prepared and measured by a Nano-Zeta Sizer at 25°C. As shown in Figure 4A, the particle size of the complexes gradually diminished on increasing the N/P ratio. The diameter of the Pluronic-PEI/DNA and PEI-SS/DNA complexes remained large (~200 nm) at even a high N/P ratio of 30 or 40, while the average particle size of Pluronic-PEI-SS/DNA complexes was smaller than 200 nm at the N/P ratio of 20. Figure 4B shows that as the N/P ratio increased, the negative charge of the complexes was almost fully neutralized and even slightly positive at the N/P ratio of 30, due to the progressive neutralization by the charged amino groups of PEI-SS segments.
As shown in Figure 5A and B, Pluronic-PEI-SS can start to form complexes at an N/P ratio of 5, but PEI-SS can complete the neutralization of DNA even at a lower N/P ratio of 3. This result demonstrates that although Pluronic shields the partially positive charges of the Pluronic-PEI-SS/DNA complex surface, the complex displays a high affinity with DNA.
We proposed that Pluronic-PEI-SS/DNA complexes containing bioreducible disulfide bonds can be kept stable in the extracellular environment, while they dissociate easily under intracellular reducing conditions due to the disulfide cleavage.23,24 To prove this proposition, the stability of the Pluronic-PEI-SS/DNA complex against nucleases was assessed by agarose gel electrophoresis to find out whether the complex can protect DNA from being degraded by nucleases. As shown in Figure 5C, Pluronic-PEI-SS/DNA complex can protect DNA from degradation even with a high concentration of DNase I (1 or 2 units/μg DNA), while the naked DNA was completely degraded by DNase I. Then, the stability of Pluronic-PEI-SS/DNA complex in the presence of DTT as cross-link sulfhydryl-reducing agent was further evaluated by gel electrophoresis assay. Figure 5D shows the effect of DTT concentration ranging from 0.5 to 310 mM on the stability of the complex with N/P ratio of 5. It was found that DNA gradually migrated with increasing concentration of DTT, suggesting the rapid release of DNA in the reducible conditions. The results suggest that the degradable Pluronic-PEI-SS complexes are capable of not only forming stable complexes with DNA from nuclease digestion but also enhancing the DNA release from the complex in cytoplasm under extreme reducing conditions.
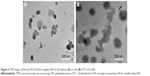
The dependence of DTT on morphology behavior of the Pluronic-PEI-SS/DNA complex was confirmed by TEM. As shown in Figure 6, the Pluronic-PEI-SS/DNA complex with a uniform size of ~80 nm at a representative N/P ratio of 20 was observed and well defined. However, the complexes swelled from 80 to 150 nm after DTT treatment. According to the GSH concentration-dependent deshielding kinetics character in the tumor tissues, the redox-responsive complex could keep stable during the circulation (5 μM GSH), slightly loosened when reaching the tumor site (20 μM GSH), and further detached from the complex in the intracellular environment (5 mM GSH). We suggest that Pluronic-PEI-SS with a suitable nanosize and a dependence on the reducible environment would facilitate the nanocarrier to enter into cells and rapidly enhance the DNA release in cytoplasm.
Cytotoxicity
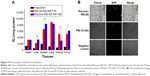
To investigate the biocompatibility of this delivery system for potential application in gene delivery, the cytotoxicity of Pluronic-PEI-SS toward HeLa and 293T cells was carried out by MTT assay, using Pluronic-PEI, PEI-SS, and PEI 25 kDa as controls. As shown in Figure 7A, the cell viability of HeLa cells treated with PEI (25 kDa) at 0.10 mg/mL was lower than 20%, indicating the high toxicity of PEI (25 kDa), whereas the cell viability after treatment with PEI-SS at the same concentration was 85% due to the biodegradable disulfide-linked low-molecular-weight PEI. Pluronic-PEI-SS and Pluronic-PEI copolymers exhibited relatively higher cell viability in comparison to PEI-SS and PEI 25 kDa. The reduction in cytotoxicity is the result of the conjugation of biocompatible Pluronic polymers and their steric shielding capable of reducing the aggregation of the positive charge on the cell surface. Especially, even at the higher concentrations used in comparison with the controls, Pluronic-PEI-SS still showed the highest cell viability, suggesting that reducible disulfide bonds and Pluronic modification attributed to good biocompatibility of the copolymer. Very similar cell viability results were also observed for the 293T cells (Figure 7B).
In vitro transfection
In vitro transfection efficiency of Pluronic-PEI-SS into HeLa and 293T cells was investigated using the pGL4 encoding EGFP-N1 (pEGFP plasmid) as reporter gene, and PEI-SS/pDNA, Pluronic-PEI/pDNA, and PEI-25 kDa/pDNA used as controls. As shown in Figure 8, the Pluronic-PEI-SS showed the highest transfection efficacy among these complexes at the N/P ratio of 30 in HeLa cells, which was approximately four, three, and even 13 times as much as that for Pluronic-PEI, PEI-SS, and 25 kDa PEI, respectively (Figure 8A). Similar results were observed in 293T cells (Figure 8B). Thus, the conjugation of Pluronic and disulfide bonding in Pluronic-PEI-SS chains could enhance the gene transfection efficiency of the HeLa and 293T cells.
Intracellular trafficking
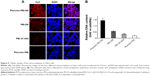
Conjugation of Pluronic with PEI-SS significantly improved both cellular uptake and transfection efficiency, possibly due to the facilitated translocation of pDNA into the nucleus by Pluronic residues and degradable PEI-SS. To prove this hypothesis, we investigated the subcellular location and delivery processes of Pluronic-PEI-SS/DNA complexes, using Pluronic-PEI, PEI-SS, and PEI 25 kDa as controls. pDNA was labeled with Cy5, and nucleus was stained with DAPI. As shown in Figure 9A, it was revealed that there was a considerably greater colocalization in the nucleus for Pluronic-PEI-SS/DNA than for the other four complexes at 4-hours post-transfection, indicating the disulfide bonding of PEI-SS and Pluronic residues could efficiently translocate Pluronic-PEI-SS/DNA into the nucleus. The ability of Pluronic-PEI-SS/DNA to enhance translocation is consistent with the previous findings.11,12
To further explore their nuclear localization, nucleocytoplasmic separation experiment was carried out to determine the concentration of pEGFP-N1 in the nuclear section (Figure 9B). HeLa cells were post-transfected with these four complexes and incubated for 4 hours, and then the cells were collected and the nuclear section was extracted. Using the nuclear section as template, the concentration of pEGFP-N1 in the nucleus was determined by real-time PCR. The results demonstrated that Pluronic-PEI-SS/DNA displayed the highest copy number of pEGFP-N1 and delivery efficiency among these nanocomplexes, which is consistent with the confocal data. In summary, efficient translocation of the Pluronic-PEI-SS/DNA complex into the nucleus may have resulted in effective transfection.
In vivo transfection
Their excellent transfection ability and low toxicity in vitro encouraged assessment of the gene expression of Pluronic-PEI-SS/DNA complex in vivo. Accordingly, male nude mice bearing HeLa cells were administered, by tail vein injection, pGL4-control reporter DNA complex. At 24-hours postinjection, mice were sacrificed to collect heart, liver, spleen, lung, kidney, and tumor. Tissue distribution of gene expression was assayed along with a corresponding nanocomplex. Compared with free naked DNA and PEI/DNA, Pluronic-PEI-SS/DNA exhibited significantly higher gene expression of luciferase in all parts of tissues. As shown in Figure 10A, gene expression of luciferase for the Pluronic-PEI-SS/DNA in liver, spleen, and tumor achieved ~3,600, 8,200, and 6,500 RLU/mg, respectively, in the organs, and higher level of luciferase activity was observed in these tissues. Especially, the transfection efficiency of Pluronic-PEI-SS/DNA in tumor was statistically significantly higher than that of PEI 25 kDa/DNA. In liver and spleen with Pluronic-PEI-SS/DNA, the transfection efficiency was statistically significantly higher than that of PEI 25 kDa/DNA complexes. The result obtained from tumor distribution also demonstrated a higher transfection efficiency of Pluronic-PEI-SS/DNA nanocomplex than that of PEI/DNA complex (P<0.05).
Furthermore, the EGFP DNA transfected cells incubated with the copolymer Pluronic-PEI-SS showed more bright green fluorescent spots than the control, as observed by fluorescence microscopy (Figure 10B). The higher in vivo transfection efficiency of the Pluronic-PEI-SS complexes can be attributed to the partial neutralization of the positive charge by Pluronic, the degradable properties of disulfide bonds, and the weak interaction between the degraded PEI short chains and DNA to enhance the release of DNA.
Conclusion
This study showed that the novel bioreducible Pluronic-PEI-SS copolymer formed by cross-linking disulfide-containing PEI (600 kDa) with Pluronic was successfully synthetized. The copolymer displayed a high affinity for DNA with a low surface charge density and suitable size, protected DNA from DNase I digestion, and also exhibited a bioreducible property in the presence of reducing agent DTT. The complex Pluronic-PEI-SS/DNA could significantly improve the in vitro transfection efficiency with low cytotoxicity, in comparison to Pluronic-PEI (25 kDa), PEI-SS, and PEI 25 kDa. Furthermore, the in vivo transfection study demonstrated that Pluronic-PEI-SS/DNA complexes induced a significantly high enrichment in the tumor and exhibited higher efficiency than the commercial PEI/DNA complex. The resulting bioreducible nanocomplex could be used as a potential gene delivery system in future.
Acknowledgments
We acknowledge the support from the National Natural Science Foundation of China (21104029, 21074049) and the Gansu Province Science Foundation for Youth (1107RJYA038). We also acknowledge the support by the grants from the Fundamental Research Funds for the Central Universities (lzujbky-2015-22) and Innovation and Entrepreneurship Program of Lanzhou University (20151073001320). We thank Prof Youqing Shen (Zhejiang University) for his help in gene delivery vectors and the animal experiment conditions.
Disclosure
The authors report no conflicts of interest in this work.
References
Cavazzana-Calvo M, Hacein-Bey S, Basile CD, et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288(5466):669–672. | ||
Yang ZR, Wang HF, Zhao J, et al. Recent developments in the use of adenoviruses and immunotoxins in cancer gene therapy. Cancer Gene Ther. 2007;14(7):599–615. | ||
Luo D, Saltzman WM. Synthetic DNA delivery systems. Nat Biotechnol. 2000;18(1):33–37. | ||
Behr JP. Synthetic gene transfer vectors II: back to the future. Acc Chem Res. 2012;45(7):980–984. | ||
Atmaja B, Lui BH, Hu YH, Beck SE, Frank CW, Cochran JR. Targeting of cancer cells using quantum dot-polypeptide hybrid assemblies that function as molecular imaging agents and carrier systems. Adv Funct Mater. 2010;20(23):4091–4097. | ||
Goyal R, Tripathi SK, Tyagi S, et al. Gellan gum blended PEI nanocomposites as gene delivery agents: evidences from in vitro and in vivo studies. Eur J Pharm Biopharm. 2011;79(1):3–14. | ||
Barua S, Joshi A, Banerjee A, et al. Parallel synthesis and screening of polymers for nonviral gene delivery. Mol Pharm. 2009;6(1):86–97. | ||
Cemazar M, Sersa G, Wilson J, et al. Effective gene transfer to solid tumors using different nonviral gene delivery techniques: electroporation, liposomes, and integrin-targeted vector. Cancer Gene Ther. 2002;9(4):399–406. | ||
Bansal R, Singh AK, Gandhi RP, Pant AB, Kumar P, Gupta KC. Galactomannan-PEI based non-viral vectors for targeted delivery of plasmid to macrophages and hepatocytes. Eur J Pharm Biopharm. 2014;87(3):461–471. | ||
Kuo JS. Effect of pluronic-block copolymers on the reduction of serum-mediated inhibition of gene transfer of polyethyleniimine-DNA complexes. Biotechnol Appl Biochem. 2003;37:267–271. | ||
Yang Z, Sahay G, Sriadibhatla S, Kabanov AV. Amphiphilic block copolymers enhance cellular uptake and nuclear entry of polyplex-delivered DNA. Bioconjuate Chem. 2008;19:1987–1994. | ||
Batrakova EV, Miller DW, Li S, Alakhov VY, Kabanov AV, Elmquist WF. Pluronic P85 enhances the delivery of digoxin to the brain: in vitro and in vivo studies. J Pharmacol Exp Ther. 2001;296(2):551–557. | ||
Lemieux P, Guerin N, Paradis G, et al. A combination of poloxamers increases gene expression of plasmid DNA in skeletal muscle. Gene Ther. 2000;7(11):986–991. | ||
Pitard B, Bello-Roufai M, Lambert O, et al. Negatively charged self-assembling DNA/poloxamine nanospheres for in vivo gene transfer. Nucleic Acids Res. 2004;32(20):e159. | ||
Gebgart LC, Sriadibhatla S, Vinogradov S, Lemieux P, Alakhov VY, Kabanov AV. Design and formulatin of polyplexes based on pluronic-polyethyleneimine conjugates for gene transfer. Bioconjugate Chem. 2002;13:937–944. | ||
Richard P, Bossard F, Desigaux L, Lanctin C, Bello-Roufai M, Pitard B. Amphiphilic block copolymers promote gene delivery in vivo to pathological skeletal muscles. Hum Gene Ther. 2005;16(11):1318–1324. | ||
Richard P, Pollard H, Lanctin C, et al. Inducible production of erythropoietin using intramuscular injection of block copolymer/DNA formulation. J Gene Med. 2005;7(1):80–86. | ||
Yang ZH, Zhu J, Sriadibhatla S, Gebhart C, Alakhov V, Kabanov A. Promoter- and strain-selective enhancement of gene expression in a mouse skeletal muscle by a polymer excipient Pluronic P85. J Control Rel. 2005;108(2–3):496–512. | ||
Liang WQ, Gong HY, Yin DF, Lu SY, Fu Q. High-molecular-weight polyethyleneimine conjuncted pluronic for gene transfer agents. Chem Pharm Bull. 2011;59(9):1094–1101. | ||
He YY, Cheng G, Xie L, Nie Y, He B, Gu ZW. Polyethyleneimine/DNA polyplexes with reduction-sensitive hyaluronic acid derivatives shielding for targeted gene delivery. Biomaterials. 2013;34(4):1235–1245. | ||
Read ML, Logan A, Seymour LW. Barriers to gene delivery using synthetic vectors. Adv Genet. 2005;53:19–46. | ||
Nguyen HK, Lemieux P, Vinogradov SV, et al. Evaluation of polyether-polyethyleneimine graft copolymers as gene transfer agents. Gene Ther. 2000;7(2):126–138. | ||
Breunig M, Lungwitz U, Liebl R, Goepferich A. Breaking up the correlation between efficacy and toxicity for nonviral gene delivery. Proc Natl Acad Sci U S A. 2007;104(36):14454–14459. | ||
Read ML, Singh S, Ahmed Z, et al. A versatile reducible polycation-based system for efficient delivery of a broad range of nucleic acids. Nucleic Acids Res. 2005;33(9):e86. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.