Back to Journals » International Journal of Nanomedicine » Volume 16
Intracellular Delivery of Proteins into Living Cells by Low-Molecular-Weight Polyethyleneimine
Authors Wu Y , jiang L, Dong Z, Chen S, Yu XY, Tang S
Received 12 April 2021
Accepted for publication 1 June 2021
Published 21 June 2021 Volume 2021:16 Pages 4197—4208
DOI https://doi.org/10.2147/IJN.S315444
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Israel (Rudi) Rubinstein
Yueheng Wu,1,2,* Lin jiang,2,* Zixuan Dong,1 Shaoxian Chen,2 Xi-Yong Yu,2 Shunqing Tang1
1National Engineering Research Center for Healthcare Devices, Guangdong Institute of Medical Instruments, Biomedical Engineering Institute, Jinan University, Guangzhou, 510632, People’s Republic of China; 2Department of Cardiovascular Surgery, Guangdong Cardiovascular Institute, Guangdong Provincial Key Laboratory of South China Structural Heart Disease, Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences, School of Medicine, South China University of Technology, Guangzhou, 510080, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Shunqing Tang
National Engineering Research Center for Healthcare Devices, Guangdong Institute of Medical Instruments, Biomedical Engineering Institute, Jinan University, Guangzhou, 510632, People’s Republic of China
Email [email protected]
Introduction: Intracellular protein delivery is emerging as a potential strategy to revolutionize therapeutics in the field of biomedicine, aiming at treating a wide range of diseases including cancer, inflammatory diseases and other oxidative stress-related disorders with high specificity. However, the current challenges and limitations are addressed to either synthetically or biologically through multipotency of engineering, such as protein modification, insufficient delivery of large-size proteins, deficiency or mutation of proteins, and high cytotoxicity.
Methods: We prepared the nanocomposites by mixing protein with PEI1200 at a certain molar ratio and demonstrated that it can deliver proteins into living cells in high efficiency and safety through the following experiments, such as dynamic light scattering, fluorescent detection, agarose gel electrophoresis, ß-Galactosidase activity detection, immunofluorescence staining, digital fluorescent detection, cell viability assay and flow cytometry.
Results: The self-assembly of PEI1200/protein nanocomposites with appropriate molar ratio (4:1 and 8:1) could provide efficiently delivery of active proteins to a variety of cell types in the presence of serum. The nanocomposites could continuously release protein up to 96 h in their desired intracellular locations. In addition, these nanocomposites were able to preserve protein activity while maintain low cytotoxicity (when final concentration < 1 μg/mL).
Conclusion: Collectively, PEI1200-based delivery system provided an alternative strategy to direct protein delivery in high efficiency and safety, offering increased potential applications in clinical biomedicine.
Keywords: polyethyleneimine, protein delivery, nanocomposites, low-molecular-weight, cytotoxicity
Introduction
Intracellular delivery of proteins is of great importance in protein-based therapeutics1,2 and biotechnologies, such as gene editing2–4 and cell reprogramming.5,6 Protein therapy protects cargo proteins against detrimental physiological environments, which exhibits higher specificity, less adverse effects, and lower risk of off-targets than gene therapy for cancer1 and genetic disorders.2 For instance, the presence of nuclease Cas9 and other genome-editing proteins avoid the sustained expression of transgenes.7 In nucleic acid-free stem cell reprogramming, direct delivery of transcriptional factor proteins can reduce the risk of insertional mutagenesis,8 which represent presumably the safest methodology for future clinical applications.
Although proteins can be delivered to living cells by protein transduction domain (PTD) carriers,9,10 cationic lipid carriers,11,12 cationic polymer13 and other molecules such as hyaluronic acid,14 most of these strategies have remained propose of disadvantages and limits. PTD carriers usually require complicated protein modification, which may cause substantial protein degradation and are prone to be captured by endosome and lysosome.9 Cationic lipid carriers (such as CRISPRMAX and PierceTM Protein Transfection Reagent) are relatively easy to use while having critical challenges due to low escape efficiency of endocytosis and phagocytosis, leading to the narrow applicability in few cell types.3,7 In addition, cationic polymer carriers are recognized as one of the most promising protein carriers, which is characterized by high capacity of delivery, wide applicability, sustained protein release and preservation of protein activity.4,15 However, the synthesis of cationic polymer needs a large amount of organic solvents and compounds mix that could significantly increase the degree of cytotoxicity and restrict the wider biomedical application.15 Therefore, it is necessary to develop an ideal platform for intracellular protein delivery with low cell toxicity as well as high expression level of protein after delivery.
Polyethyleneimine (25 kDa, PEI 25k) generally has the extensive capacity to deliver not only nucleic acids16 but also multiple proteins into living cells,17,18 while challenges remain in clinical application due to high cytotoxicity and poor biodegradability. A common strategy for reducing toxicity is to graft other biological materials with low-molecular-weight PEI.19,20 For example, researchers previously synthesized polymers by grafting various guanidyl containing compounds with low-molecular-weight PEI, which theoretically enabled efficient delivery and maintenance of bioactivity for various cargo proteins.19 However, these conventional grafting methods are subject to unstable performance of protein delivery due to non-standard experimental procedures,21 and the low density of low-molecular-weight PEI.22
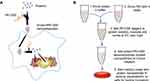
Given the potential applicable strengths and challenges presented, we reported a protein delivery method based on self-assembly of low-molecular-weight PEI (1.2 kDa, PEI1200) and proteins into stable nanocomposites (hereinafter, the protein-PEI1200 nanocomposites are denoted as ‘protein nanoparticles’) via charge interactions (Figure 1A and B). The proteins were transduced into living cells and protected from rapid degradation by extracellular and endosomal proteases. To date, low-molecular-weight PEI based is a prominently novel tool to achieve intracellular delivery of proteins of interest, with notable potentials in future biomedical application.
Materials and Methods
Proteins
GFP (Sino Biological, China), BSA (Thermo Scientific, USA), DNase I (ComWin Biotech, China) and β-galactosidase (Sigma-Aldrich, Germany) were purchased from referred companies. HAND2 proteins were contributed from Dr. Xiaohong Li in Guangdong Provincial People’s Hospital, which has been described in previous publications.23,24 For the detailed properties of the proteins used in this study, please see Table S1.
PEI1200 Reagent
A stock solution of PEI1200 reagent was prepared with 100 mg of PEI with a molecular weight of 1200 (Sigma-Aldrich) and 10 mL of PBS solution (Gibco), and the final stock concentration of the PEI1200 reagent was 10 mg/mL.
Preparation of Protein Nanoparticles Using PEI1200 Reagent
The proteins were diluted in PBS solution or deionized water, and the stock concentration was approximately 1 mg/mL by BCA protein assay. The PEI1200 reagent was titrated into the protein solution dropwise at 4°C. Stir the mixture of PEI1200 reagent and proteins gently and slowly to facilitate the self-assembly reaction of protein-PEI1200 nanocomposites (protein nanoparticles) at a variety of molar ratio. The final concentration of protein nanoparticles was so-called 1 mg/mL. The final pH was adjusted to 7.4.
Delivery of Protein Nanoparticles to Living Cells
Four types of cells were involved in our study. Human breast adenocarcinoma MCF-7 cell line (MCF-7, ATCC), human dermal fibroblasts (HDFs, Gibco) and human umbilical vein endothelial cells (HUVEC, ATCC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Normocin, Invitrogen) at 37°C in a fully humidified 5% CO2 atmosphere. Uniformly, 5 mL culture medium with different concentration of protein nanocomposites was added into 1×106 cells. For hBMSCs, cells were obtained from the bone marrow of a patient (This study was approved by the Institutional Review Board at Guangdong General Hospital, No. GDREC2016255H, has been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki, and the participant signed an informed consent form prior to research commencement). Mononuclear cells were isolated using a Histopaque density gradient of 1077 g/mL. Primary hBMSCs culture was performed as described previously.25 The passaged hBMSCs were cultured in OriCell™ medium (Cyagen, China) until the number of cells was sufficient for protein delivery, which occurred between passage 3 and 6. The identification of hBMSCs was performed by flow cytometry as our previous publication.25
The protein nanoparticles were diluted with cell culture medium, and the working concentration of delivered protein depends on each experimental purpose. The delivery efficiency was detected by microscopy, immunofluorescence staining or digital fluorescent detection. The positive control was performed using PierceTM Protein Transfection Reagent (Thermo Scientific, USA), following the manufacturer’s instructions, which is a commercially available cationic reagent.
Fluorescent Detection of GFP in hBMSCs
The hBMSCs were cultured in 35-mm cell dishes (Falcon, USA) at the density of 60%, transduced by GFP nanoparticles with a molar ratio of 16:1 to 2:1 (PEI1200/GFP) for 24 h. Nuclei were stained with Hoechst 33342 (Roche, Switzerland). Fluorescent images of living cells were acquired using a confocal microscope (TCS-SP5, Leica). For alternative protocol, cells were incubated with ER Tracker Red (Thermo Fisher, USA) for 30 min following the manufacturer’s instructions.
Particle Size, Zeta Potential and TEM Morphology of Protein Nanoparticles
Both particle size and zeta potential were measured on dynamic light scattering (DLS) by Zetasizer Nano ZS (Malvern, v.7.13) with a He-Ne laser (λ = 632 nm). In order to examine its stability in the culture medium, the DNase I nanoparticles were dissolved in DMEM with or without 10% FBS. The control groups are PBS solution and deionized water. The concentration of DNase I nanoparticles in culture medium was 0.5, 1, 2 and 4 μg/mL, respectively.
For the transmission electron microscopy (TEM) detection, 5 μL of DNase I nanoparticles solution was dropped onto carbon TEM grids, deposited for 1 min before blotting with filter paper, then air-dried overnight. The TEM grids were imaged under microscopy (Philips Tecnai 10, Netherlands). DNase I nanoparticles solution was diluted in PBS at 50 μg/mL.
Encapsulation Efficiency of Protein Nanoparticles
To obtain the protein encapsulation efficiency, protein nanoparticles were prepared as working condition of 1 mg/mL in aqueous suspension at pH 7.4, and separated by 13,500 rpm centrifugation for 15 min. The supernatant was collected to evaluate the residual protein by BCA protein assay kit (Pierce, Thermo Fisher, USA). Encapsulation efficiency was calculated indirectly as follows. Encapsulation efficiency = ((Initial protein concentrations – Unencapsulated protein concentrations)/Initial protein concentrations) × 100%.
Agarose Gel Electrophoresis (AGE)
The reversible inactivation effect of DNase I was detected by AGE. PEI1200/DNase I complexes (50 µg/mL) or DNase I (50 µg/mL) were mixed with Cmyc plasmid (1.6 µg/mL) in 30 min at room temperature. The mixture was electrophoresed in 1% agarose gel with 1 × DNA loading buffer for 30 min and observed under UV light.
ß-Galactosidase Activity Detection
HDFs and hBMSCs were transduced with 5 μg/mL ß-galactosidase nanoparticles incubated in the culture medium for 24 h. Cells were fixed for 10 min on ice with 0.2% glutaraldehyde (Polysciences, USA) in PBS. Rinse in detergent solution (0.02% Igepal, 0.01% sodium deoxycholate in PBS); then, incubated overnight at 37°C in detergent solution containing 1 mg/mL X-gal (Thermo Fisher, USA) plus 5 mM potassium ferricyanide and 5 mM potassium ferrocyanide, shielded from light.
Immunofluorescence Staining
Cells were fixed in 4% paraformaldehyde (Sigma-Aldrich, Germany) for 10 min, and permeabilized in PBS containing 0.1% Triton X-100 (Sigma-Aldrich) for 10 min. After treatment with 5% BSA blocking for 1 h at room temperature, the primary antibody incubations were performed overnight at 4°C followed by the appropriate Alexa Fluor-conjugated secondary antibodies (Invitrogen, USA) for 2 h at room temperature, and nuclei were stained with Hoechst 33342 (Roche, Switzerland). Images were acquired using a confocal microscope (TCS-SP5, Leica).
Digital Fluorescent Detection
Cells were fixed and permeabilized as previously description, blocked with Odyssey® Infrared Imaging System Blocking Buffer (LI-COR, Lincoln, NE, USA) for 1 h, followed by incubate with primary antibody in Odyssey blocking buffer at 4°C overnight. After washed with PBS containing 0.1% Tween (PBST), samples were incubated with goat anti-rabbit antibody (Abcam) conjugated with IRDye® 800CW (LI-COR) at room temperature for 1 h. Fluorescence was detected using an Odyssey CLx Infrared Imaging System (LI-COR), and signal intensity was quantified with Image Studio™ Lite (Version 3.1, LI-COR) following software instructions.
Cell Viability Assay (MTT)
MTT assay was used to determine the cell viability of hBMSCs following manufacturer’s instruction. Cells were seeded in 96-well plate at 1×104/well density for 24 h, incubated with HAND2 nanoparticles in different concentrations. After 72 h, assays were performed by adding 50 μL of MTT solution (5 mg/mL MTT formazan, Sigma-Aldrich). The culture plate then incubated for an additional 4 h at 37°C, 5% CO2, and 95% humidity. After removal of MTT solution, the crystals formed at the bottom of the culture plate were solubilized in 150 μL of acid isopropyl alcohol, recording the absorbance at 570 nm with an automatic microplate reader (Multiskan GO, Thermo Fisher, USA). The quantity of formazan product as measured is directly proportional to the cell viability. The experiment was repeated by three times.
Flow Cytometry
Approximately 1×106 of hBMSCs in each treatment were harvested with 0.05% trypsin-EDTA digestion, fixed on ice with 4% paraformaldehyde for 10 min. The single-cell suspension was centrifuged at 800 rpm for 5 min before discarding the supernatant and repeating the procedure. Staining was done in PBS containing 0.2% BSA and Annexin V-PI (Sigma-Aldrich) used followed the manufacturer’s recommendation. Blank control (non-staining) and single-positive control (Annexin V-FITC staining or PI staining) were set to access the compensation of double channels. Data were collected using a flow cytometer (FACSAria II, USA) and analyzed via CellQuest software. The apoptosis rate = (FITC+/PI+ number of cells + FITC+/PI− number of cells)/total number of cells × 100%.
Statistical Analyses
Statistical analysis was performed using GraphPad Prism 10 (GraphPad Software, Inc). The significance of the difference between two groups was determined by an unpaired Student’s t-test. All graphic data were presented as mean±SD, P-value < 0.05 was considered statistically significant.
Results
Screening for Optimal Composited Molar Ratio of PEI1200/Protein
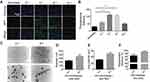
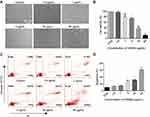
In order to screen for the optimal PEI1200/protein molar ratio, we tested 4 groups of PEI1200-DNase I complexes (PEI1200/DNase I molar ratio at 16:1, 8:1, 4:1 to 2:1, respectively). First, we examined the efficiency of GFP protein delivery into hBMSCs and found that fluorescence label at PEI1200/protein ratio of 16:1 or 2:1 was obviously lower than that at 8:1 or 4:1 (Figure 2A). Meanwhile, we measured the accurate fluorescence intensity of intracellular GFP that released from the GFP nanoparticles via Odyssey CLx Infrared Imaging System. And the peak of fluorescence was observed at the ratio of 4:1 (Figure 2B).
To confirm the formation of PEI1200/protein nanocomposites, we investigated the morphology, zeta potential and particle size of PEI1200/DNase I complex at different molar ratios (16:1, 8:1, 4:1 to 2:1, respectively). The protein-PEI1200 nanocomposites were uniformly dispersed (Figure 2C-E) at the molar ratio of 8:1 (with average diameter of 359 nm and zeta potential of +45 mV) and 4:1 (with average diameter of 244 nm and zeta potential of +25 mV). In contrast, stable nanocomposites could not be synthesized at the ratio of 16:1 or 2:1 (Figure S1a, S1b). Similarly, when the molar ratio of PEI1200 and other proteins (such as GFP, BSA, HAND2 and β-galactosidase) is 4:1, stable nanocomposites can also be formed (Table S1). These results showed that when PEI1200 and proteins are self-assembled in an appropriate molar ratio, nanocomposites with relatively uniform size can be obtained. The PEI1200/DNase I nanocomposites in a range of nanoparticle concentrations (0.5 μg/mL to 4 μg/mL) were also uniformly dispersed in DMEM with or without 10% FBS, and similar with deionized water or PBS solution (Figure S2a, S2b).
Considering the non-covalent property of PEI1200-protein binding, we focused on the investigation of nanocomposite encapsulation efficiency at pH 7.4. When the PEI1200/BSA molar ratio at 8:1 and 4:1, the encapsulation efficiency was 98.3% and 91.9%, respectively (Figure 2F), which indicated the stability of PEI1200/BSA complex during the transduction. Thus, we found that the encapsulation efficiency of PEI1200/BSA ratio at 8:1 is higher than 4:1, and the PEI1200/BSA ratio of 8:1 is more stable than 4:1, which is consistent with the laser scattering experiment (as described above). Similarly, the encapsulation efficiency of other PEI1200/protein (GFP, DNase I, HAND2 and β-galactosidase) nanocomposites is about 90% at the PEI1200/protein ratio of 4:1 (Table S1).
Maintenance Potency of Protein Biological Activity in Living Cells
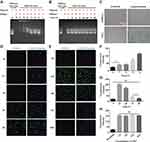
Besides the membrane impermeability, protein can be easily degraded and inactivated in response to serum in the culture medium, and further evidence of cytosolic access was demonstrated through fast captured and inactivated by lysosomes and endosomes in living cells. Here we designed a series of experiments to evaluate the activity of protein nanoparticles before and post transduction. Firstly, we examined a series of complexes with a ratio of PEI1200/DNase I ranging from 128:1 to 2:1 to measure degradation of Cmyc plasmid driven by DNase I via agarose gel electrophoresis. When the ratio of PEI1200/protein was 128:1 to 4:1, DNase I was temporarily inactivated and could not act on Cmyc plasmid (Figure 3A). When the ratio of PEI/protein was 2:1, Cmyc plasmid was almost completely degraded by DNase I. This result indicated that DNase I nanoparticles could temporarily block biological function of degrading plasmids within a wide range of PEI1200/protein ratio, which is consistent with the protection effect given by PEI1200. Time course of plasmid degradation suggested that DNase I nanoparticles was persistently inactivated and prevented to act on Cmyc plasmid from 1 h to 96 h (Figure 3B), which further corroborated the stability of the PEI1200/protein nanocomposites.
In order to evaluate the activity of proteins delivered into cells, β-galactosidase was used to demonstrate functional delivery of an active and biomedical enzyme. The β-galactosidase was self-assembled with PEI1200 and delivered into HDFs and hBMSCs, whose biological activity was assessed by the color change when response to substrate X-gal.26,27 The results (Figure 3C) suggested that although enzymes were susceptible to inactivation during the delivery process, they retained activity inside the cells. Furthermore, we also investigated the time-course uptake of HAND2 nanoparticles in living hBMSCs by tracing the metabolism of FITC signal. Fluorescence labeling showed that few proteins had been delivered into cells starting at 1 h, while the peak of fluorescence intensity was seen from 8 h to 24 h. And signal disappeared in approximately 96 h at the same concentration (Figure 3D-G). This finding indicated that HAND2 nanoparticles could efficiently enter living cells and perform post-uptake release of HAND2 protein, leading to ideal transient metabolism without accumulation in the cell.
Notably, the essential presence of serum in the cell culture medium significantly inhibits the cell uptaking of protein nanoparticles28,29 due to a variety of enzymes involved.30,31 However, conventional serum-free methods not only increase the complexity of the experiment design, but also affects cell growth and protein delivery efficiency. In contrast, HAND2 nanoparticles were delivered into hBMSCs at serum concentrations of 1%, 5%, 10%, and 20% in our study, showing that the serum culture does not affect the HAND2 delivery efficacy (Figure 3H).
Protein Nanoparticles Stably Delivered into Various Cell Lines in High Efficiency and Indicative of Intracellular Localization
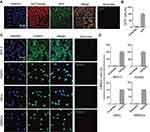
Given the above evidences of PEI1200-mediated delivery of GFP, β-galactosidase, and HAND2 to hBMSCs or HDFs through protein nanoparticles, it remains unclear whether the delivered proteins are distributed throughout the cell with identical cellular localization according to different cell types and protein properties. Therefore, we focused on two proteins (GFP and HAND2) in four cell lines (HDFs, hBMSCs, MCF-7 and HUVEC). GFP was evenly distributed throughout the cytoplasm of hBMSCs with almost 100% expression as compared to scramble group. And the fluorescence was partially co-localized with the endoplasmic reticulum tracking system (Figure 4A and B), which suggested gradually post-uptake release. Remarkably, as a transcription factor (TF), delivered HAND2 were mainly enriched in the nucleus of cells regardless of cell types, since DNA binding domains in TFs are accessible to bind to targeted genes in the nucleus (Figure 4C). Meanwhile, immunostaining indicated that >97% of cells were positive for delivered HAND2 in four cell types, further demonstrated self-assembled protein nanoparticles were much more highly transduced into a variety of cells as compared with control methods (Figure 4D). However, commercial protein transfection reagent such as PierceTM showed no capacity to achieve stem cell protein delivery yet (Figure S3). Moreover, neither GFP nor HAND2 was enriched in the lysosomes or endosomes of all cell types, indicating that protein nanoparticles could escape from the capture by endosomes and lysosomes. Interestingly, we found HAND2 were also partially located in the cytoplasm of MCF-7 cells (Figure 4C). That might be because cytoplasm of cancer cells contains considerable extrachromosomal circular DNA32,33 and extrachromosomal circular DNA, which processes regions bind to TF protein. In summary, this method can deliver not only proteins with large molecular weight (135KD, β-galactosidase), but also negatively charged (GFP, β-galactosidase) and positively charged (HAND2) proteins into living cells (Table S1). This result showed the high transportation efficacy in PEI1200-mediated protein delivery, as well as the flexibility of targeted cell type and protein properties.
Induction of Cytotoxicity in hBMSCs by Protein Nanoparticles
Considerably, the transient cell permeabilization provided by these natively cationic nanoparticles allowed influx of numerous proteins into cells, inevitably generating potential cytotoxicity and side effects. Thus, we examined the effects of protein nanoparticles on PEI-induced cytotoxicity via cell viability and Annexin V-PI apoptosis assays. We found that treatment of hBMSCs with HAND2 nanoparticles for 72 h resulted in dose-dependent induction of cytotoxicity at the concentration of 1/4-64 μg/mL. Therefore, we compare the proliferative capacity of protein nanoparticles treated cells and blank control at different concentrations. According to the MTT measurement, the cell viability of 1/4 μg/mL and 1 μg/mL group slightly decreased by 5–9% after HAND2 nanoparticles treatment (P>0. 05), while the proliferative viability of hBMSCs significantly decreased after exposure to 4 μg/mL treatment (P<0. 05), and further increasing the concentration of HAND2 nanoparticles would lead to the precipitation of cytotoxicity (P<0. 01) (Figure 5B). Moreover, typical shrinkage and weakness in hBMSCs were observed when concentration was over 4 μg/mL, accompanied with lower proliferative capacity (Figure 5A). Likewise, to clarify the mechanism of nano-induced apoptosis and cell death in hBMSCs, flow cytometry was used to investigate both early and late apoptosis using Annexin V-FITC and PI labeling of living cells. In our study, it is shown that protein nanoparticles notably simulate early apoptosis (increase ~4%) and late apoptosis (increase ~14%) for hBMSCs at 4 μg/mL treatment compared to blank control. And this induced apoptosis would gradually deteriorate till approximately 60% of cells were suffering the side effect in 64 μg/mL treatment (Figure 5C and D). However, be of interest as neither of treatment group seemed to driven from late apoptosis to necrosis. Conclusively, the results suggested that dose-dependent cytotoxicity in hBMSCs was induced by protein nanoparticles after incubation for 72 h, speeding up the decline in cell cycle. So, we select 1 μg/mL protein nanoparticles treatment for subsequent exploration to facilitate delivery efficiency and minimize side effects.
Discussion
In this study, based on the PEI1200/protein self-assembly into nanocomposites, we developed a convenient solution to protein delivery into living cells. It only requires to mix the protein and PEI1200 in a certain ratio to deliver the protein into living cells. To our knowledge, this method represents the most convenient application of low-molecular-weight PEI to date that enables delivery of active proteins with low cytotoxicity.
It is well known that high-molecular-weight PEI can condense nucleic acids into nanometers,34 which is not the case for low-molecular-weight PEI.35,36 The low-molecular-weight PEI can be compounded with high-molecular materials to prepare gene carriers, which are usually low in toxicity and can deliver nucleic acids into living cells with relatively low efficiency. However, considering the fact that protein is characterized by a denser structure than nucleic acid, we assumed that the carriers are not needed to condense proteins. And low-molecular-weight PEI with low charge density may spontaneously interact with proteins to form self-assembled nanocomposites. The experimental results of this study strongly supported our hypothesis.
The results of fluorescent detection showed that our method is not interfered by the serum in medium. This may be because the cationic nature of protein nanoparticles temporarily suppresses the activity of delivered proteins, thus preventing their interactions with the enzymes in serum. In addition, the proteins delivered into cells are highly active. This may be due to the non-covalent binding of protein and PEI1200. Unlike some protein delivery methods that require protein modification,10,37,38 our strategy does not destroy the protein structure.
Protein delivery into nucleus is of great importance for a variety of biotechnologies, such as gene editing. Although cationic lipid is used to deliver cas9 protein into nucleus, the formation of protein-lipid complexes requires sophisticated fusion to either a highly negatively charged protein such as (–30) GFP or complexation with polyanionic nucleic acids.7 Polymeric nanoparticle-based delivery to the cell nucleus also needs complicated tagging of nuclear localization sequences (NLS).39 There were also studies on the delivery of transcription factors by polymers. For example, deliver OCT4 into the nucleus of Sf9 cells by chitosan.5 The highly complex and sophisticated operation hindered the broader application of the above techniques. In contrast, the method described in the present study is one of the most convenient approaches for delivering proteins into cell nucleus.
The cytotoxicity of PEI40,41 is mainly attributed to cell necrosis caused by the destruction of the cell membrane and cell apoptosis induced by the rupture of the mitochondrial membrane.42,43 Even though low-molecular-weight PEI is less toxic, it remains necessary to identify the critical concentration of toxicity, in order to guarantee the biomedical applications in cell reprogramming and gene editing. We observed no toxic effect on the proliferation and apoptosis at a low concentration of protein nanoparticles (<1 μg/mL). Our previous results suggested that the protein concentration of 0.5 μg/mL is practical in cell reprogramming within the range of PEI1200-based delivery.24 Therefore, the method presented in this study should also be applicable in gene editing, which only requires a relatively low amount of protein to be delivered.4,11 For example, using modified liposome technology to deliver 3ng/mL cas9 protein, the target gene can be edited.12
In summary, we developed a protein delivery method based on PEI1200. We simply mixed PEI1200 and protein at a ratio of 4:1 and 8:1 to obtain nanocomposites, which can enter living cells with high concentration of serum. We also demonstrated that our method can deliver various proteins to multiple cell types without destroying protein activity. In broad biomedical applications, this method may serve as a more convenient and cost-efficient strategy for protein delivery than other alternatives.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Liu X, Wu F, Ji Y, Yin L. Recent advances in anti-cancer protein/peptide delivery. Bioconjugate Chemi. 2019;30(2):305–324. doi:10.1021/acs.bioconjchem.8b00750
2. Gao X, Tao Y, Lamas V, et al. Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature. 2018;553(7687):217–221.
3. Wang M, Zuris JA, Meng F, et al. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc Natl Acad Sci USA. 2016;113(11):2868–2873.
4. Chen G, Abdeen AA, Wang Y, et al. A biodegradable nanocapsule delivers a Cas9 ribonucleoprotein complex for in vivo genome editing. Nat Nanotechnol. 2019;14(10):974–980.
5. Tammam S, Malak P, Correa D, et al. Nuclear delivery of recombinant OCT4 by chitosan nanoparticles for transgene-free generation of protein-induced pluripotent stem cells. Oncotarget. 2016;7(25):37728–37739.
6. Khan M, Narayanan K, Lu H, et al. Delivery of reprogramming factors into fibroblasts for generation of non-genetic induced pluripotent stem cells using a cationic bolaamphiphile as a non-viral vector. Biomaterials. 2013;34(21):5336–5343.
7. Zuris JA, Thompson DB, Shu Y, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2014.
8. Chou MJ, Yu HY, Hsia JC, et al. Highly efficient intracellular protein delivery by cationic polyethyleneimine-modified gelatin nanoparticles. Materials (Basel). 2018;11:2.
9. Kristensen M, Birch D, Morck Nielsen H. Applications and challenges for use of cell-penetrating peptides as delivery vectors for peptide and protein cargos. Int J Mol Sci. 2016;17:2.
10. Salerno JC, Ngwa VM, Nowak SJ, Chrestensen CA, Healey AN, McMurry JL. Novel cell-penetrating peptide-adaptors effect intracellular delivery and endosomal escape of protein cargos. J Cell Sci. 2016;129(12):893–897. doi:10.1242/jcs.192666
11. Kim YB, Zhao KT, Thompson DB, Liu DR. An anionic human protein mediates cationic liposome delivery of genome editing proteins into mammalian cells. Nat Commun. 2019;10(1):2905.
12. Zuris JA, Thompson DB, Shu Y, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33(1):73–80.
13. Galliani M, Tremolanti C, Signore G. Nanocarriers for protein delivery to the cytosol: assessing the endosomal escape of poly(Lactide-co-Glycolide)-Poly(Ethylene Imine) nanoparticles. Nanomater (Basel). 2019;9:4.
14. Li JA, Chen L, Zhang XQ, Guan SK. Enhancing biocompatibility and corrosion resistance of biodegradable Mg-Zn-Y-Nd alloy by preparing PDA/HA coating for potential application of cardiovascular biomaterials. Mater Sci Eng C Mater Biol Appl. 2020;109:110607.
15. Villegas MR, Baeza A, Vallet-Regi M. Nanotechnological strategies for protein delivery. Molecules. 2018;23:5.
16. Lemkine GF, Demeneix BA. Polyethylenimines for in vivo gene delivery. Current Opinion Mol Ther. 2001;3(2):178–182.
17. Futami J, Kitazoe M, Maeda T, et al. Intracellular delivery of proteins into mammalian living cells by polyethylenimine-cationization. J Biosci Bioeng. 2005;99(2):95–103.
18. Murata H, Futami J, Kitazoe M, et al. Intracellular delivery of glutathione S-transferase-fused proteins into mammalian cells by polyethylenimine-glutathione conjugates. J Biochem. 2008;144(4):447–455.
19. Zhao J, Yang L, Huang P, et al. Synthesis and characterization of low molecular weight polyethyleneimine-terminated Poly(beta-amino ester) for highly efficient gene delivery of minicircle DNA. J Coll Interface Sci. 2016;463:93–98.
20. Liu YK, Lin YL, Chen CH, et al. A unique and potent protein binding nature of liposome containing polyethylenimine and polyethylene glycol: a nondisplaceable property. Biotechnol Bioeng. 2011;108(6):1318–1327.
21. Lv J, Tan E, Wang Y, Fan Q, Yu J, Cheng Y. Tailoring guanidyl-rich polymers for efficient cytosolic protein delivery. J Controlled Release. 2020;320:412–420.
22. Breunig M, Lungwitz U, Liebl R, et al. Gene delivery with low molecular weight linear polyethylenimines. J Gene Med. 2005;7(10):1287–1298.
23. Sivashanmugam A, Murray V, Cui C, Zhang Y, Wang J, Li Q. Practical protocols for production of very high yields of recombinant proteins using Escherichia coli. Protein Sci2009;18(5):936–948.
24. Li XH, Li Q, Jiang L, et al. Generation of functional human cardiac progenitor cells by high-efficiency protein transduction. Stem Cells Transl Med. 2015;4(12):1415–1424.
25. Huang W, Xiao DZ, Wang Y, et al. Fn14 promotes differentiation of human mesenchymal stem cells into heart valvular interstitial cells by phenotypic characterization. J Cell Physiol. 2014;229(5):580–587.
26. Meka AK, Abbaraju PL, Song H, et al. A vesicle supra-assembly approach to synthesize amine-functionalized hollow dendritic mesoporous silica nanospheres for protein delivery. Small. 2016.
27. Ayame H, Morimoto N, Akiyoshi K. Self-assembled cationic nanogels for intracellular protein delivery. Bioconjugate Chemi. 2008;19(4):882–890.
28. Drake DM, Keswani RK, Pack DW. Effect of serum on transfection by polyethylenimine/virus-like particle hybrid gene delivery vectors. Pharmaceutical Res. 2010;27(11):2457–2465.
29. Kumari M, Liu CH, Wu WC. Efficient gene delivery by oligochitosan conjugated serum albumin: facile synthesis, polyplex stability, and transfection. Carbohydrate Polymers. 2018;183:37–49.
30. van der Gun BT, Monami A, Laarmann S, et al. Serum insensitive, intranuclear protein delivery by the multipurpose cationic lipid SAINT-2. J Controlled Release. 2007;123(3):228–238.
31. Sabaa MW, Hanna DH, Abu Elella MH, Mohamed RR. Encapsulation of bovine serum albumin within novel xanthan gum based hydrogel for protein delivery. Materi Sci Eng C Mater Biol Appl. 2019;94:1044–1055.
32. Sin STK, Jiang P, Deng J, et al. Identification and characterization of extrachromosomal circular DNA in maternal plasma. Proc Natl Acad Sci USA. 2020;117(3):1658–1665.
33. Chiu RWK, Dutta A, Hensson AG, Lo YMD, Mischel P, What RB. Is extrachromosomal circular DNA and what does it do? Clin Chem. 2020;66(6):754–759.
34. Ge X, Feng J, Chen S, et al. Biscarbamate cross-linked low molecular weight Polyethylenimine polycation as an efficient intra-cellular delivery cargo for cancer therapy. J Nanobiotechnol. 2014;12:13.
35. Zeng J, Wang X, Wang S. Self-assembled ternary complexes of plasmid DNA, low molecular weight polyethylenimine and targeting peptide for nonviral gene delivery into neurons. Biomaterials. 2007;28(7):1443–1451.
36. Wen Y, Pan S, Luo X, Zhang X, Zhang W, Feng M. A biodegradable low molecular weight polyethylenimine derivative as low toxicity and efficient gene vector. Bioconjugate Chem. 2009;20(2):322–332.
37. Jo J, Hong S, Choi WY, Lee DR. Cell-penetrating peptide (CPP)-conjugated proteins is an efficient tool for manipulation of human mesenchymal stromal cells. Sci Rep. 2014;4:4378.
38. Hou Y, Protein LH. PEPylation: a new paradigm of protein-polymer conjugation. Bioconjugate Chem. 2019;30(6):1604–1616.
39. Tammam SN, Azzazy HM, Breitinger HG, Lamprecht A. Chitosan nanoparticles for nuclear targeting: the effect of nanoparticle size and nuclear localization sequence density. Mol Pharm. 2015;12(12):4277–4289.
40. Xiong MP, Forrest ML, Ton G, Zhao A, Davies NM, Kwon GS. Poly(aspartate-g-PEI800), a polyethylenimine analogue of low toxicity and high transfection efficiency for gene delivery. Biomaterials. 2007;28(32):4889–4900.
41. Bauer JA, Rao W, Smith DJ. Evaluation of linear polyethyleneimine/nitric oxide adduct on wound repair: therapy versus toxicity. Wound Repair Regeneration. 1998;6(6):569–577.
42. Hunter AC. Molecular hurdles in polyfectin design and mechanistic background to polycation induced cytotoxicity. Advan Drug Del Rev. 2006;58(14):1523–1531.
43. Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol Ther. 2005;11(6):990–995.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.