Back to Journals » OncoTargets and Therapy » Volume 9
Integrating subpathway analysis to identify candidate agents for hepatocellular carcinoma
Authors Wang J, Li M, Wang Y, Liu X
Received 26 September 2015
Accepted for publication 23 December 2015
Published 7 March 2016 Volume 2016:9 Pages 1221—1230
DOI https://doi.org/10.2147/OTT.S97211
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr William C. Cho
Jiye Wang,1 Mi Li,2 Yun Wang,3 Xiaoping Liu4
1The Criminal Science and Technology Department, Zhejiang Police College, Hangzhou, Zhejiang Province, 2Department of Nursing, Shandong College of Traditional Chinese Medicine College, Yantai, Shandong Province, 3Office Department of Gastroenterology, The First Affiliated Hospital of Xi’an Jiao Tong University, Xi’an, Shanxi Province, 4Key Laboratory of Systems Biology, Shanghai Institutes for Biological Sciences, Shanghai, People’s Republic of China
Abstract: Hepatocellular carcinoma (HCC) is the second most common cause of cancer-associated death worldwide, characterized by a high invasiveness and resistance to normal anticancer treatments. The need to develop new therapeutic agents for HCC is urgent. Here, we developed a bioinformatics method to identify potential novel drugs for HCC by integrating HCC-related and drug-affected subpathways. By using the RNA-seq data from the TCGA (The Cancer Genome Atlas) database, we first identified 1,763 differentially expressed genes between HCC and normal samples. Next, we identified 104 significant HCC-related subpathways. We also identified the subpathways associated with small molecular drugs in the CMap database. Finally, by integrating HCC-related and drug-affected subpathways, we identified 40 novel small molecular drugs capable of targeting these HCC-involved subpathways. In addition to previously reported agents (ie, calmidazolium), our method also identified potentially novel agents for targeting HCC. We experimentally verified that one of these novel agents, prenylamine, induced HCC cell apoptosis using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, an acridine orange/ethidium bromide stain, and electron microscopy. In addition, we found that prenylamine not only affected several classic apoptosis-related proteins, including Bax, Bcl-2, and cytochrome c, but also increased caspase-3 activity. These candidate small molecular drugs identified by us may provide insights into novel therapeutic approaches for HCC.
Keywords: subpathways, hepatocellular carcinoma, small molecules, network
Introduction
Hepatocellular carcinoma (HCC), the sixth most common cancer worldwide, accounts for 95% of primary liver cancer.1 HCC is the second most common cause of cancer-associated death worldwide and was estimated to be responsible for ~746,000 deaths in 2012.2,3 If diagnosed early, HCC can be treated curatively with surgical resection or liver transplantation; however, since the majority of HCC patients are diagnosed at an advanced stage, their median survival times are generally <1 year, leading to a poor prognosis.4,5 Additionally, very few patients are candidates for liver resection, and liver donors are generally uncommon.2 Furthermore, other modalities such as radiofrequency ablation and transarterial chemoembolization have poor efficacy.2,6 Therefore, special and effective chemotherapy for treating HCC is still needed.
However, the development of novel anticancer drugs is time consuming and risky. Therefore, drug repurposing, identifying new indications for known drugs, has become an effective and innovative approach. One strategy for drug repurposing is to distinguish and investigate pathways related to HCC development and then identify potential agents that may affect these pathways. As bioinformatics databases and methods have developed, high-throughput screening of HCC has become possible, providing global cancer genome information. One example is The Cancer Genome Atlas (TCGA) database, which is a community resource project, in which data are made available rapidly after generation for community research use. Additionally, databases allowing the investigation of gene signatures after small molecule treatments, such as the CMap database, have been constructed to provide microarray data under drug-induced conditions.7
To explore new agents for HCC, we implemented a bioinformatics method for drug repurposing of HCC based on integrating subpathway analyses. The differentially expressed genes (DEGs) between HCC tissues and nontumor tissues were identified from the TCGA database. Then, we identified HCC-related pathways. After treatment with small molecules in CMap, affected pathways were also identified. Finally, small molecule drugs that are capable of targeting these HCC-related subpathways were considered potential novel agents for HCC treatment. We experimentally verified that prenylamine, one of the potential agents we identified, could induce apoptosis of HepG2 (HCC cell line). Finally, we explored the apoptotic mechanisms of prenylamine by measuring the expression of apoptosis-related proteins, including Bax, Bcl-2, cytochrome c, and caspase-3 activity. In brief, the candidate agents identified by our approach may provide important insights in improving the therapeutic status of HCC cases.
Materials and methods
Microarray data and DEG analysis
We downloaded RNA-seq data from liver HCC samples from the TCGA database (http://cancergenome.nih.gov/). These data are based on human HCC tissues isolated from normal and HCC patients. A total of 371 HCC samples and 49 normal samples were sequenced on the Illumina Genome Analyzer RNA Sequencing platform. We downloaded the TCGA level 3 data for further analysis. We used edgeR, a Bioconductor software package for examining differential expression of replicated count data,8 to identify DEGs. The genes with a fold-change value >2 or <0.5 and with a false discovery rate (FDR) <0.01 were considered DEGs.
Identifying subpathways by SubpathwayMiner
HCC-related subpathways were identified using SubpathwayMiner an R-based software package that flexibly identifies subpathways using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway structure information database.9 KEGG (http://www.genome.jp/kegg/) is a collection of online databases containing information on genomes, enzymatic pathways, and biological chemicals. In the SubpathwayMiner package, the pathway structure data of KEGG are converted to undirected R graph objects, in which enzymes are considered nodes; and if the two enzymes participate in the same reactions, two nodes are connected by an edge. To identify subpathways, in which all enzymes have similar functions, from entire pathways, the k-clique concept in social network analysis was adopted. This software can define subpathways based on distance similarity among enzymes.9 In social network analysis, a k-clique in a graph is defined as a subgraph, in which the distance between any two nodes is not greater than a parameter, specified as k.9 Here, we set k=3, a setting at which the distance among enzymes in one subpathway is not >3.
We then imported the DEGs in HCC into SubpathwayMiner to identify significantly enriched subpathways. An FDR <0.1 was considered statistically significant.
Global associations between drugs and metabolic subpathways
First, we identified the global association between 1,309 bioactive small molecular agents from the CMap database and the identified subpathways from the SubpathwayMiner.7 We downloaded microarray profiles for each agent from the CMap database and obtained the affected genes for each small molecule. The CMap database contains 6,100 instances corresponding to 1,309 bioactive small molecules. Each instance includes treatment and control gene expression profiles, corresponding to treatment with or without a bioactive small molecule administered to the cells. We downloaded all the microarray profiles and their associated annotation files from the CMap website (http://www.broadinstitute.org/cmap/). For each instance, we matched perturbation and control pairs of expression profiles according to descriptions of the instances in the annotation file. Then, we used fold-change analysis to identify DEGs, for each instance with a fold change >2 or <0.5, between the corresponding treatment and control gene expression profiles. The DEGs were merged if the corresponding instances (experiments) belonged to the same bioactive small molecule, and these genes were considered to be affected genes for this agent.
Next, we entered the affected genes for each small molecule into SubpathwayMiner to identify significantly enriched subpathways for each small molecule. An FDR <0.1 was chosen as the cutoff for statistically significant subpathways.
Identification of novel drugs for HCC
To find novel agents, which may play a role in HCC-involved subpathways, we integrated the aforementioned global associations between drugs and subpathways and HCC-involved subpathways. If drug-related subpathways were also involved in HCC, the corresponding drugs were considered potential drugs for the treatment of HCC.
HepG2 cell culture
The human hematoma HepG2 cell line was purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, People’s Republic of China) and cultured in Dulbecco’s Modified Eagle’s Medium containing 10% (v/v) heat-inactivated fetal bovine serum at 37°C in an incubator containing humidified air with 5% (v/v) CO2. No ethics statement was required from the institutional review board for the use of these cell lines.
Cell viability assay
Cells were seeded onto 96-well plates at 8×103 cells per well 24 hours before treatment. The cells were treated with the prenylamine (at 0, 10, 50, 100, 200, and 500 μmol/L). After 12 hours, 15 μL (5 mg/mL) 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)10 (Sigma-Aldrich Co., St Louis, MO, USA) was added to each well, followed by incubation at 37°C for 4 hours. Then, the MTT solution was removed, and 150 μL dimethyl sulfoxide was added. The mixtures were shaken for 10 minutes to fully dissolve the crystals. A microplate reader (Tecan) was used to measure the absorbance at a wavelength of 570 nm. Cell viability was expressed as percentage changes in the absorbance values from the treatment group over the control group.
Electron microscopy
HepG2 cells were cultured in 60 mm plates, collected in phosphate-buffered saline (PBS) solution, and fixed with 2% (v/v) paraformaldehyde containing 2.5% (w/v) glutaraldehyde (Paesel & Lorei) buffered in Hank’s modified salt solution at 4°C for 4 hours. The cells were further fixed in 1% (w/v) OsO4 solution buffered by 0.1 M cacodylate (pH 7.2) at 4°C for 2 hours, after which the cells were scraped off from the plastic and dehydrated in ethanol. Dehydration was completed in propylene oxide. The specimens were embedded in Araldite (Serva). Ultrathin sections were produced on an FCR Reichert Ultracut ultra microtome (Leica Microsystems), mounted on pioloform-coated copper grids, and contrasted with lead citrate. Specimens were analyzed and documented with an EM 10A electron microscope (Carl Zeiss Meditec AG).
Acridine orange/ethidium bromide fluorescence staining
HepG2 cells in the exponential growth phase were cultivated on sterile coverslips for 24 hours and were subsequently treated with 200 μmol/L for 12 hours. The cells were washed twice with PBS and then mixed with 1 mL of dye mixture containing 100 mg/mL acridine orange (AO) and 100 mg/mL ethidium bromide (EB) in PBS.11 Cellular morphological changes were examined using fluorescence microscopy (×200). The percentage of apoptotic cells was calculated by the following formula: apoptotic rate (%) = number of apoptotic cells/number of all cells counted.12,13
Cell apoptosis assays
Double staining with Annexin V-FITC and propidium iodide (BD, Franklin Lakes, NJ, USA) was performed for quantification of apoptosis of HepG2 cells. Both the attached cells and the cells in the supernatant were collected, washed twice with ice-cold PBS, and resuspended in 400 mL of binding buffer. Annexin V-FITC and propidium iodide were added to the cells and incubated for 15 minutes in the dark at 4°C. After incubation, the samples were analyzed by a flow cytometry (BD FACSCalibur; BD), and 2.0×104 events per sample were counted.
Western blotting
Total protein was extracted from the HepG2 cells for immunoblotting analysis. Protein samples (80 μg of protein) were separated in a 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis gel and blotted onto nitrocellulose membranes. After blotting, the membranes were probed with Bax, Bcl-2, cytochrome c, and glyceraldehyde-3-phosphate dehydrogenase antibodies and incubated overnight at 4°C. Infrared fluorescent dye-labeled secondary antibody was incubated with the membrane for 1 hour. Western blot bands were collected using the infrared Imaging System (LI-COR Biosciences), and the band density was quantified using Odyssey 3.0 software for each group and normalized against glyceraldehyde-3-phosphate dehydrogenase.
Caspase-3 activity assay
Caspase-3 activity was analyzed using a caspase-3 activity assay kit (Beyotime Institute of Biotechnology, Shanghai, People’s Republic of China) according to the manufacturer’s instructions, using the substrate peptides Ac-DEVD-p-nitroanilide (pNA), Ac-IETD-pNA, and Ac-LEHD-pNA. Briefly, the supernatant of the cell lysate was mixed with buffer containing the substrate peptides for caspase attached to pNA. The release of pNA was quantified by determining the absorbance with an enzyme-linked immunosorbent assay reader at 405 nm. Caspase activities were expressed as percentage over control.
Result
DEGs between HCC cases and healthy controls
In order to explore the mechanisms underlying HCC, we obtained the expression profiles of HCC samples and normal samples from the TCGA database. We identified a total of 1,763 genes as differentially expressed between HCC patients and controls (“Materials and methods” section).
Subpathway enrichment analysis
Using SubpathwayMiner with a k value set to 3, we identified 104 enriched subpathways corresponding to 42 entire pathways with an FDR <0.1 (Tables 1 and S1). These 104 metabolic subpathways and the corresponding entire pathways were considered significantly related to the development of HCC. Furthermore, we identified global associations between 1,309 bioactive small molecular agents from the CMap database and these subpathways. Using a cutoff of FDR <0.01, a total of 840 associations between 196 small molecules and the 104 subpathways were obtained (Table S2).
Drug repurposing for HCC
To identify candidate agents that are capable of targeting HCC-related subpathways, we integrated the aforementioned global associations between drugs and metabolic subpathways (“Materials and methods” section).14 By integrating these 104 HCC-related subpathways and small molecules that affected these subpathways, we selected the overlapping subpathways that are related to both HCC and small molecules. Thus, we assumed that the small molecules, which affected the overlapping subpathway, might also play a role in perturbing the development of HCC. There were eight overlapping subpathways corresponding to a total of 40 small molecules (Table 2).
  | Table 2 Potential novel drugs and their information |
We then built a network between these repositioned drugs and the overlapping metabolic subpathways by integrating the relationships mentioned earlier (Figure 1). In this network, the rectangle nodes represent drugs and the circle nodes represent subpathways. The subpathways are color coded by the entire pathway in which they participate (Figure 1).
Of 40 small molecules, prenylamine was chosen for next validation not only because of its safety and known mode of action but also due to its apoptosis potential. It was always used as antianginal drug for decades.15 This class of medicaments mimicked the cardiac effects of simple Ca2+ withdrawal in that they diminished Ca2+-dependent high energy phosphate utilization, contractile force, and oxygen requirement of the beating heart without impairing the Na-dependent action potential parameters.15 Furthermore, several studies had reported the pivotal role of Ca2+ homeostasis in cell apoptosis.16 Therefore, we next chose prenylamine to treat HepG2 cells.
Prenylamine suppresses the viability of HepG2 cells
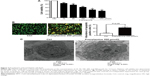
The antiproliferative effect of prenylamine on HepG2 cells was examined by exposing the cells to various concentrations (0, 10, 50, 100, 200, and 500 μmol/L) of prenylamine for 12, 24, and 48 hours (Figure S1). We found that cell growth was inhibited in a dose-dependent manner (Figure 2A). In the presence of 200 μmol/L prenylamine, HepG2 cells exhibited ~50% inhibition of proliferation after treatment for 12 hours. Hence, this concentration and a 12-hour treatment time were used in the following experiments.
Prenylamine induces apoptosis in HepG2 cells
To investigate whether prenylamine regulates apoptosis, AO/EB staining and electron microscopy were used to detect the apoptotic cells. The results from our fluorescence microscopy analysis are shown in Figure 2B. Prenylamine induced substantial cell apoptosis (P<0.05). Under an electron microscope, the cells treated with prenylamine exhibited robust changes in microstructure, including cell surface microvillus reduction, nuclear chromatin condensation, margination, and membrane blistering10 (Figure 2C). In addition, apoptotic body was found (the white arrow in Figure 2C).
We also determined whether prenylamine affected HCC cell apoptosis using a flow cytometry. After being treated with prenylamine for 12 hours, the late apoptosis of HepG2 cells was higher than the control group (Figure S2A and B). These data implied that prenylamine induces apoptosis of HCC cells.
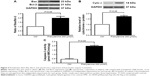
Prenylamine activates proapoptotic signaling pathways
To explore the mechanisms by which prenylamine induced apoptosis in HepG2 cells, we measured the expression of downstream proteins in apoptotic pathways, including Bax, Bcl-2, and cytochrome c. The results demonstrated that treatment with prenylamine could upregulate Bax and cytochrome c and downregulate Bcl-2 expression (Figure 3A and B). In addition, caspase-3 activity, as a marker of apoptosis, was increased (1.9-fold) by prenylamine (Figure 3C).
Discussion
Analysis of gene expression in disease can reveal the underlying gene activity changes, which contribute to the disease and can pinpoint potential targets for improving therapeutic intervention. In this study, RNA-seq data from the TCGA database were used to explore the mechanisms of HCC development. A total of 1,763 DEGs were identified between HCC samples and healthy controls. The subpathway mining results showed 104 metabolic subpathways corresponding to 42 entire pathways that were dysfunctional in HCC. Then, we identified potential therapeutic agents for HCC using a computational drug repurposing method. A total of 40 small molecules were identified that are potential drug candidates for perturbing the development of HCC.
The majority of current pathways enrichment approaches commonly focus on entire pathways. However, a previous study showed that subpathway analysis may be more suitable than entire pathways for the identification of drug responses.9 In this study, we identified 104 subpathways among 42 KEGG pathways. Some of these subpathways are well documented as involved in the development of HCC. For example, the top three significant subpathways belong to the extracellular matrix–receptor interaction pathway (path: 04512; P=3.54×10−11; FDR =8.16×10−8; Tables 1 and S1), which consists of molecules such as CD36, CD44, and CD104 that serve an important role in the direct or indirect control of cellular activities such as adhesion, migration, differentiation, proliferation, and apoptosis.17 Molecules in this pathway have been shown to drive pathological angiogenesis and tumor progression.18 The cell cycle pathway (path: 04110_17; P=5.59×10−05; FDR =0.003), referring to the reproducible sequence of events in the G1 and G2 phases of the cell cycle, was also detected. Key regulatory enzymes of this pathway are cyclin-dependent kinases, which are dysregulated and serve as potential therapeutic targets in HCC.19,20 Additionally, we also identified other cancer-related subpathways, such as the peroxisome proliferator-activated receptor signaling pathway (path: 03320) and pathways in cancer (path: 05200).
We then identified a group of known drugs with potential novel therapeutic efficacy for HCC by integrating the association between small molecules and subpathways. A study by Li et al also identified global associations between 403 metabolic subpathways and 488 known drugs using SubpathwayMiner with k=3.21 However, in that study, only metabolic subpathways were investigated rather than all kinds of pathways.21 Some signaling pathways also play an important role in drug response. Therefore, in our study, we implemented subpathway enrichment to all kinds of pathways. Of 104 drug-affected subpathways, eight subpathways were also involved in the development of HCC. These overlapping subpathways corresponded to 40 small molecules.
Some of these small molecules were found to have apoptotic effects. For example, calmidazolium induced apoptosis of human hepatoma cells,22 and niclosamide inhibited proliferation as well as inducing apoptosis and cell cycle arrest in human osteosarcoma cell lines.23 However, the effects of some other drugs are not well characterized. Prenylamine, a calcium antagonist, has been used for the treatment of angina pectoris.24 Although some early studies have investigated its effects on a colon cancer cell line,14 there have been no direct reports about the apoptotic effects of prenylamine on HCC cells. To further investigate the effects of prenylamine on HCC, we treated HepG2 cells with different concentrations (10, 50, 100, 200, and 500 μmol/L) of prenylamine. The MTT assay indicated that prenylamine could induce HepG2 cell apoptosis in a slightly dose-dependent manner. AO/EB staining and electron microscopy also detected the apoptotic cells. Finally, we found that prenylamine not only affected some classic apoptosis-related proteins, including Bax, Bcl-2, and cytochrome c, but also increased caspase-3 activity, suggesting that prenylamine might regulate multiple potential targets, directly or indirectly influencing these proteins. Although our approach based on targeting subpathways successfully identified potential agents for HCC, further experiments are needed to explore the mechanisms underlying these observations and to ensure the safety of these small molecules for use in HCC clinical treatment.
Disclosure
The authors alone are responsible for the content and writing of the paper. The authors report no conflicts of interest in this work.
References
Deng GL, Zeng S, Shen H. Chemotherapy and target therapy for hepatocellular carcinoma: new advances and challenges. World J Hepatol. 2015;7(5):787–798. | ||
Hong YP, Li ZD, Prasoon P, Zhang Q. Immunotherapy for hepatocellular carcinoma: from basic research to clinical use. World J Hepatol. 2015;7(7):980–992. | ||
Lau WY. Primary liver tumors. Semin Surg Oncol. 2000;19(2):135–144. | ||
Bruix J, Sherman M; American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53(3):1020–1022. | ||
Giakoustidis A, Giakoustidis D, Mudan S, Sklavos A, Williams R. Molecular signalling in hepatocellular carcinoma: role of and crosstalk among WNT/ss-catenin, Sonic Hedgehog, Notch and Dickkopf-1. Can J Gastroenterol Hepatol. 2015;29(4):209–217. | ||
Pez F, Lopez A, Kim M, Wands JR, Caron de Fromentel C, Merle P. Wnt signaling and hepatocarcinogenesis: molecular targets for the development of innovative anticancer drugs. J Hepatol. 2013;59(5):1107–1117. | ||
Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. | ||
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. | ||
Li C, Li X, Miao Y, et al. SubpathwayMiner: a software package for flexible identification of pathways. Nucleic Acids Res. 2009;37(19):e131. | ||
Zheng F, He K, Li X, et al. Transient overexpression of TGFBR3 induces apoptosis in human nasopharyngeal carcinoma CNE-2Z cells. Biosci Rep. 2013;33(2):e00029. | ||
McGahon AJ, Martin SJ, Bissonnette RP, et al. The end of the (cell) line: methods for the study of apoptosis in vitro. Methods Cell Biol. 1995;46:153–185. | ||
Chen H, Takahashi S, Imamura M, et al. Earthworm fibrinolytic enzyme: anti-tumor activity on human hepatoma cells in vitro and in vivo. Chin Med J. 2007;120(10):898–904. | ||
Ribble D, Goldstein NB, Norris DA, Shellman YG. A simple technique for quantifying apoptosis in 96-well plates. BMC Biotechnol. 2005;5:12. | ||
Batra S, Alenfall J. Effect of diverse categories of drugs on human colon tumour cell proliferation. Anticancer Res. 1991;11(3):1221–1224. | ||
Fleckenstein A. History of calcium antagonists. Circ Res. 1983;52(2 pt 2):I3–I16. | ||
Zhang S, Chen Y, Wu X, et al. The pivotal role of Ca homeostasis in PBDE-47-induced neuronal apoptosis. Mol Neurobiol. Epub 2015 Dec 17. | ||
Nistico P, Di Modugno F, Spada S, Bissell MJ. beta1 and beta4 integrins: from breast development to clinical practice. Breast Cancer Res. 2014;16(5):459. | ||
Giancotti FG. Targeting integrin beta4 for cancer and anti-angiogenic therapy. Trends Pharmacol Sci. 2007;28(10):506–511. | ||
Li W, Long J, Takikawa Y, Suzuki K, Lin S. Plasma from patients with acute liver failure dampens HepG2 cells to epidermal growth factor induced proliferation response. Hepatogastroenterology. 2014;61(135):2021–2027. | ||
Zhao H, Bauzon F, Bi E, et al. Substituting threonine 187 with alanine in p27Kip1 prevents pituitary tumorigenesis by two-hit loss of Rb1 and enhances humoral immunity in old age. J Biol Chem. 2015;290(9): 5797–5809. | ||
Li C, Shang D, Wang Y, et al. Characterizing the network of drugs and their affected metabolic subpathways. PLoS One. 2012;7(10): e47326. | ||
Liao WC, Huang CC, Cheng HH, et al. Effect of calmidazolium on [Ca2+]i and viability in human hepatoma cells. Arch Toxicol. 2009;83(1):61–68. | ||
Li Z, Yu Y, Sun S, Wang W, Yu A. Niclosamide inhibits the proliferation of human osteosarcoma cell lines by inducing apoptosis and cell cycle arrest. Oncol Rep. 2015;33(4):1763–1768. | ||
Beuck S, Sigmund G, Koch A, et al. Identification and characterization of urinary prenylamine metabolites by means of liquid chromatography-tandem mass spectrometry. Drug Test Anal. 2012;4(9):701–716. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.