Back to Journals » Journal of Inflammation Research » Volume 15
Integrated Analysis of Ferroptosis and Immunity-Related Genes Associated with Intestinal Ischemia/Reperfusion Injury
Authors Zhu L , Lian W, Yao Z, Yang X, Wang Z, Lai Y, Xu S , Zhao B, Liu
Received 8 December 2021
Accepted for publication 1 April 2022
Published 14 April 2022 Volume 2022:15 Pages 2397—2411
DOI https://doi.org/10.2147/JIR.S351990
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Adam D Bachstetter
Lin Zhu,* Wanyi Lian,* Zhiwen Yao, Xiao Yang, Ziyi Wang, Yupei Lai, Shiting Xu, Bingcheng Zhao, Kexuan Liu
Department of Anesthesiology, Nanfang Hospital, Southern Medical University, Guangzhou, Guangdong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Kexuan Liu; Bingcheng Zhao, Department of Anesthesiology, Nanfang Hospital, Southern Medical University, Guangzhou, Guangdong, People’s Republic of China, Tel/Fax +86 020 61641881, Email [email protected]; [email protected]
Purpose: Intestinal ischemia/reperfusion (I/R) injury is an unresolved clinical challenge due to its high prevalence, difficulty in diagnosis, and lack of clinically effective therapeutic agents. Ferroptosis is a novel form of cell-regulated death that has been shown to play a role in various I/R models and has been shown to be immune-related. Further unraveling the molecular mechanisms associated with ferroptosis and immunity in intestinal I/R injury may lead to the discovery of potentially effective drugs.
Methods: We obtained differentially expressed mRNAs (DEGs) in mouse intestinal tissues following intestinal I/R injury or sham surgery. Then, we extracted ferroptosis-related DEGs (FRGs) and immune-related DEGs (IRGs) from the DEGs. In addition, we performed functional analysis of FRGs and IRGs. Next, we used transcriptome sequencing from patients with intestinal I/R injury to validate the results. Then, we constructed transcription factors (TFs)-gene networks and gene-drug networks using mouse and human co-expressed FRGs (coFRG) and mouse and human co-expressed IRGs (coIRG). We also analyzed the composition of immune cells to reveal correlations between FRGs signatures and immune cells in the mouse and human gut. Finally, we validated these results through animal experiments.
Results: We extracted 61 FRGs and 294 IRGs from mouse samples and performed PPI and functional analyses. We extracted 45 FRGs and 200 IRGs from human samples for validation, and identified 24 coFRGs,100 coIRGs and 6 hub genes (HSPA5, GDF15, TNFAIP3, HMOX1, CXCL2 and IL6) in both. We also predicted potential TF-gene networks for coFRGs and coIRGs, as well as predicted gene-drug pairs for hub genes. In addition, we found that the immune cells were altered in the early stages of intestinal I/R injury and that FRGs were closely associated with immune cells in mice and humans. Finally, we validated the hub genes in mouse samples.
Conclusion: In conclusion, we identified ferroptosis and immunity-related genes to predict their correlations in intestinal I/R injury. We also predicated potential TF-genes network and potential therapeutic targets (HSPA5, GDF15, TNFAIP3, HMOX1, CXCL2 and IL6) to provide clues for further investigation of intestinal I/R injury.
Keywords: intestinal ischemia-reperfusion injury, ferroptosis, immunity, RNA-seq, bioinformatics
Introduction
Intestinal ischemia/reperfusion (I/R) injury is a common life-threatening pathophysiological condition that can cause harmful complications, including systemic inflammatory responses and multi-organ dysfunction syndromes.
I/R injury has been studied in various organs and several key concepts regarding its mechanisms have been unraveled, but the exact molecular mechanisms and pathways remain obscure and still need to be investigated.1 Therefore, exploring the mechanisms of intestinal I/R injury and finding drugs to alleviate the damage has become a therapeutic priority.
Ferroptosis, a novel type of cell-regulated death caused by iron-dependent lipid peroxidation, has been implicated in a variety of pathophysiological processes and is a potential therapeutic target for the treatment of many diseases.2 Accumulating evidence suggests that ferroptosis is crucially involved in the mechanism for cell death occurred during I/R injury in different organs, including brain, kidney and intestine.3–7 Due to temporary blood flow deprivation and restoration, intracellular redox homeostasis is disrupted after I/R process. Ferroptosis is caused by intracellular lipid peroxidation and is highly dependent on the intracellular redox state and iron availability. Deng et al demonstrated that overexpression of Gpx4 inhibited ferroptosis induced by intestinal I/R and Li et al showed that activation of ACSL4 contributed to ferroptosis-mediated tissue damage in intestinal I/R.5,7 However, as a complex biological process, ferroptosis is regulated by several genes, mainly involving genetic changes in iron homeostasis and lipid peroxidation metabolism, but the specific regulatory mechanisms need further studied.8
In addition, the role of ferroptosis in immunotherapy has aroused researcher’s interest recently.9 Proneth’s10 review demonstrated that some physiological processes induced by ferroptosis could activate innate immune system in early phases to some extent. Meanwhile, O’Donnell et al elucidated that enzymatically oxidized phospholipids formed in immune cells have roles in ferroptosis.11 However, the precise molecular mechanism and pathway of ferroptosis-immunity crosstalk associated with intestinal I/R injury is not well understood. What is more, Yan et al12 summarized in the review that iron chelation alleviated ischemia-reperfusion injury in many animal models, but its use was limited in the clinical setting due to efficiency of action, side effects, etc. Therefore, it is vital to have further insight into reliable biomarkers and therapeutic targets of ferroptosis-immune crosstalk in intestinal I/R injury, and that is the purpose of this article.
With the development of high-throughput technology, sequencing technology can be used to detect the genes that differ in intestinal I/R injury in batches, thus improving the reliability and accuracy of research. This study is the first integrated analysis of differentially expressed mRNAs (DEGs) of ferroptosis and immunity after intestinal I/R injury, as well as the first analysis of immune cell infiltration and its correlation with ferroptosis after intestinal I/R. Our study provides research ideas and potential therapeutic targets for the treatment of ferroptosis and immune disorders associated with intestinal I/R injury.
Materials and Methods
Experimental Design
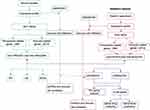
The experimental design of the study was shown in Figure 1. DEGs between mouse intestinal I/R group and sham group (mmu-DEGs) which were analyzed from mouse transcription sequencing results were intersected separately with Ferroptosis-related gene sets and immune-related gene sets which were obtained separately from the database FerrDb and ImmPort to obtain the Ferroptosis-related DEGs (FRGs) and immune-related DEGs (IRGs). The FRGs from mouse were named mmu-FRGs, and the IRGs from mouse were named mmu-IRGs. mmu-FRGs and mmu-IRG were uploaded to the STRING database for protein–protein interaction (PPI) analysis. Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed on mmu-FRGs and mmu-IRGs, respectively.
The transcriptome sequencing data of intestinal I/R group and the control group in human were downloaded from the European Nucleotide Archive (ENA) of EMBL-EBI to verify the results in mouse. First, DEGs from human samples (hsa-DEGs) were analyzed, and then the FRGs from humans (hsa-FRGs) and the IRGs from humans (hsa-IRGs) were obtained as above. The intersection of mmu-FRGs and hsa-FRGs was the FRGs co-expressed in mouse and human (coFRGs), and the same as the IRGs co-expressed in mouse and human (coIRGs). CoFRGs and coIRGs were uploaded to NetworkAnalyst to obtain Transcription Factors (TFs) paired with coFRGs and coIRGs. The intersection of coFRGs and coIRGs were the hub genes related to both ferroptosis and immunity, and these 6 hub genes were uploaded to the DGIdb database to obtain related drugs that might be potential targets for intestinal I/R damage.
In addition, the expression profile of mouse and human were upload to CIBERSORT separately to obtain the immune cell infiltration matrix after intestinal I/R in both mouse and human separately. The correlation between coFRGs and infiltrating immune cells in both mouse and human were analyzed by Spearman correlation analysis. Finally, the expression of these 6 hub genes and other related indicators were verified in the mouse model.
Data
Intestinal samples derived from male C57BL/6 (weight 20–25g, 7–8w) mice subjected to intestinal I/R (n = 4) or sham surgery (n = 4) were collected and the mRNA expression profiles were obtained from our previous RNA sequencing analysis accession number PRJNA784411 (https://www.ebi.ac.uk/ena/browser/view/PRJNA784411). Small intestine expression profiles of 5 intestinal I/R injury patients (group A) and 5 controls (group C) were downloaded from the ENA at EMBL-EBI under accession number PRJEB45362 (https://www.ebi.ac.uk/ena/browser/view/PRJEB45362).13
Ferroptosis-related genes were obtained from the database FerrDb (http://www.zhounan.org/ferrdb/), which is the world’s first database for ferroptosis, providing an updated database for regulators and markers as well as ferroptosis-disease associations, and were identified by searching and reading original articles from PubMed, and 390 ferroptosis-related genes were obtained finally.14
A list of 2013 immunity-related genes was obtained (Updated: July 2020) from the Immunology Database and Analysis Portal (ImmPort, https://www.immport.org/home), which was a database that updated immunology data accurately and in a timely manner. Data from ImmPort were a powerful foundation of immunology research. These genes were proven to participate in the process of immune activity.
Differentially Expressed Genes Analysis
The data from mice were divided into two groups: the intestinal I/R group (CIR) and the Sham operated group (CS), and the data from patients were divided into two groups: group A and group C. The DESeq2 package (version 1.30.1) was used to identify the DEGs between CIR and CS. DEGs were defined as p value < 0.05 and a |log2FoldChange| > 1. The pheatmap package (version 1.0.12) was applied for the bidirectional hierarchical clustering, and the expression values were presented using heatmaps.15
PPI Network Generation and Module Analysis
The STRING database (version:11.5, https://string-db.org) was used to construct PPI network for the mmu-FRGs and mmu-IRGs encoded proteins, including direct and indirect associations. The Required Confidence (combined score) > 0.7 was set as threshold value. The Cytoscape (version 3.8.2) was utilized to visualize the raw data of network from STRING database and then displayed.16
GO and KEGG Analysis of FRGs and IRGs
GO enrichment analysis and KEGG were performed in R software with clusterProfiler package and was visualized by ggplot2 package (3.3.5) and enrichplot package in R Studio.17
TFs Matching Prediction
The coFRGs and coIRGs were uploaded to NetworkAnalyst (https://www.networkanalyst.ca/) to obtain the TFs paired with coFRGs and coIRGs. These TFs were predicted from the ENCODE database and used to generate a list of TFs-genes pairs outcomes. The results were visualized in Cytoscape.
Drugs Prediction
The hub genes which appeared in both coFRGs and coIRGs serve as candidate targets to find therapeutic drugs in the DGIdb database (http://dgidb.genome.wustl.edu/). The discovery of genes-drug interactions was also visualized in Cytoscape, and the filter of DGIdb was set to “FDA approved.”
Genemania
Genemania database (https://genemania.org/) was used to analyze protein and genetic interactions, co-expression, co-localization, pathways and protein domain similarity of the hub genes.18
Evaluation of Immune Cell Infiltration
We uploaded the gene expression profile data from mouse and human separately to CIBERSORT (https://cibersort.stanford.edu/), which is an analytical tool to provide an estimation of the abundance of member cell types in a mixed cell population, using gene expression data,19 and to obtain the immune cell infiltration matrix. Then, we used ggplot2 package to draw boxplot to visualize the differences in immune cell infiltration.
Correlation Analysis Between FRGs and Infiltrating Immune Cells
The Hmisc package (4.5.0) was used to perform Spearman correlation analysis on coFRGs and infiltrating immune cells from both mice and humans and corrplot package was used to visualize the results.
Validation of Ferroptosis Related DEGs in Mouse Intestinal I/R Injury Model
Animal Model
Twelve male C57BL/6 (weighing 20–25g, 7–8w) mice were obtained from Nanfang Hospital, Southern Medical University, divided into two groups randomly (CIR, intestinal I/R injury group; CS, sham operated group) and then housed at separated cages in a temperature-controlled room under a fixed circadian rhythm with free access to food and water. The animal experiments were approved by the Ethics Committee of Nanfang Hospital (China, application No: NFYY-2021-0267) and all procedures were carried out in compliance with National Institutes of Health guidelines for the use of experimental animals. According to a previous study, the intestinal I/R injury was generated by occluding the superior mesenteric artery (SMA) with a microvascular clamp for 60 minutes followed by 240 minutes of reperfusion.7,18,19 In addition, the Sham group was the same with intestinal I/R injury except occluding the SMA. Outcome evaluators and analysts were blinded to the treatment to which the mice were assigned.
Sample Collection and Histological Staining
Intestine segments were collected at the end of reperfusion and then were divided into three segments, two of the segments were dried on filter paper and preserved at −80°C, waiting for detection, and one segment was treated in 4% paraformaldehyde as previously described.21 Images were captured at 200× and 400× with BX63 automated fluorescence microscope (Olympus, Tokyo, Japan). The degree of intestinal mucosal damage after reperfusion was assessed using Chiu’s scoring system22 by two independent blinded technicians.
Glutathione (GSH) Assays
Serum GSH concentrations were detected by the GSH Assay Kit (Abcam, ab239709) following the manufacturer’s instructions.
Western Blotting
The concentration of protein extracted from mouse intestinal tissue samples was determined by Western blotting. Briefly, the gut tissues were lysed by prechilled RIPA lysis buffer containing with protease and phosphatase inhibitor cocktail and PMSF for 10 minutes. The protein was quantified using BCA method, and equal amount of them were separated on 15% SDS PAGE followed by transferred to NC membranes. The membranes were blocked in 5% non-fat milk for 1.5 hours and then incubated with primary antibodies overnight: rabbit anti-Ferritin Heavy Chain (Cat. ab183781, Abcam, dilution 1:1000), anti-Gpx4 (Cat. ab125066, Abcam, dilution 1:1000), anti-ACTB (Cat. AP0060, Bioworld, 1:5000). Then the membrane was washed by TBST (0.1% Tween 20) and incubated with secondary antibody (Cat.RM3002, Beijing Ray Antibody Biotech, 1:10000). The ECL detection reagent was used to visualize the protein bands in the membrane. The quantity of the bands was analyzed with ImageJ.
Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR) Analysis
Trizol reagent was used to extract total RNA according to the manufacturer’s protocol. The mRNAs were reversed into cDNA by kits from Toyobo, according to the previous study.20 mRNA quantitative analysis was conducted with qRT-PCR using Toyobo SYBR Green Realtime PCR (Applied Biosystems, Foster City, CA) .18S was served as reference gene, and the fold change relative to the control was computed by the 2−ΔΔCt method. Table 1 displays the Forward and Reverse primer sequences.
 |
Table 1 The Primer Sequences of qRT-PCR |
Statistical Analysis
The statistical software package SPSS26.0 and R (version 4.0.3) were used for statistical computation. The experimental results were checked with the Shapiro–Wilk test for normality and Levene test for homogeneity of variance. Normally distributed quantitative data were expressed as mean ± standard error mean and Student’s t-tests were utilized for the analysis between two groups. Data with an abnormal distribution were expressed as median and differences were compared using the Mann–Whitney U-test. Correlation analysis was calculated using the Spearman’s coefficient (r), and with r > 0.6 and p < 0.05 considered significant. Two-sided p < 0.05 was considered statistically significant (*p < 0.05, **p < 0.01, ***p < 0.001).
Results
Identification of Ferroptosis and Immunity Related Genes
We obtained 3571 mmu-DEGs from 4 intestinal I/R injury samples and 4 sham-operated samples from mouse in this experiment (Table S1, Figure S1). Then, we extracted 61 mmu-FRGs from 390 entries of the FerrDb database and literature reporting and 294 mmu-IRGs from 2013 entries of the ImmPort database (Figure 1, Figure 2D–E, Figures S2 and 3, Tables S2 and 3).
Identification of coFRGs and coIRGs
To make the results clinically applicable, we obtained 2245 hsa-DEGs from five patients with arterial ischemic intestinal I/R injury and five patients without intestinal I/R injury (Table S4, Figure S4A and B).
Then, we extracted 45 hsa-FRGs and 200 hsa-IRGs from human samples (Figure 2F–G, Figures S5 and 6, Tables S5 and 6). CoFRGs were FRGs that were expressed in both mouse and human and are identical to Co-FRGs. Finally, 24 coFRGs and 100 coIRGs were identified (Figure 2A and B, 2H and I, Figures S7 and 8). Of these, 6 hub genes (including HSPA5, GDF15, TNFAIP3, HMOX1, CXCL2 and IL6) were present in both coFRGs and coIRGs (Figure 2C).
Construction Protein–Protein Interaction Network of the FRGs and IRGs
The mmu-FRGs and mmu-IRGs expressed in mouse were constructed into a PPI network by STRING database, and visualized by Cytoscape (Figure 3A). The network contained 284 nodes and 1312 protein-pairs.
Functional Enrichment Analyses
We performed GO and KEGG functional enrichment analysis on these 61 mmu-FRGs and 294 mmu-IRGs to investigate the potential function of the ferroptosis-related genes in mice.
The GO analysis showed that the mmu-FRGs were mainly related to the response to oxidative stress, positive regulation of cellular catabolic process, regulation of apoptotic signaling pathway, neuro death, autophagy and process utilizing autophagic mechanism for biological processes (BPs), apical part of cell, lipid droplet and basolateral plasma membrane for molecular functions (MFs), and organic anion transmembrane transporter activity, ubiquitin protein ligase binding and ubiquitin-like protein ligase binding for cellular components (CCs). The mmu-IRGs were mostly enriched in the response to cytokine-mediated signaling pathway, positive regulation of response to external stimulus, regulation of inflammatory response, cellular calcium ion homeostasis, leukocyte migration for BPs, receptor complex, plasma membrane signaling receptor complex, endoplasmic reticulum lumen for MFs, and receptor ligand activity, cytokine receptor binding for CCs (Figure 3B–D, Tables S7 and 8).
KEGG enrichment analysis demonstrated that mmu-FRGs were mostly enriched in the ferroptosis, autophagy-animal, IL-17 signaling pathway, TNF signaling pathway, HIF-1 signaling pathway, glutathione metabolism, and the IRGs were mostly enriched in the cytokine-cytokine receptor interaction, neuroactive ligand-receptor interaction, IL-17 signaling pathway, TNF signaling pathway, lipid and atherosclerosis, etc. (Figure 3C–E, Tables S9 and 10).
Establishment of TF-Genes Network and Genes-Drugs Network
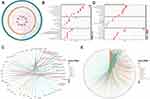
To explore more underlying functions of coFRGs and coIRGs, TF-genes and genes-drugs relationships were further analyzed. In TF-genes analysis, there were 55 nodes and 141 edges in TF-coFRGs pairs (Figure 4A), and 91 nodes and 406 edges in TF-coIRGs pairs (Figure 4B). To limit the scope, the 6 hub genes were used for further investigation. In genes-drugs network analysis, 6 genes and 31 drugs were identified (Figure 4C).
Genemania
There are protein-protein interactions within the 6 hub genes. The protein and genetic interactions, co-expression, co-localization, pathways and protein domain similarity of the hub genes are shown in Figure 4D–I.
Evaluation of Immune Cell Infiltration
We uploaded the gene expression profile data from mouse and human separately to CIBERSORT, and obtained the immune cell infiltration matrix. As shown in Figure 5A, immune cell including macrophages M0, eosinophils and activated memory CD4 T cells were significantly increased intestinal I/R injury in mice. Meanwhile, macrophages M1 was increased and macrophages M2 was decreased in intestinal I/R injury.
In human, immune cells including neutrophils and activated mast cells were significantly different in intestinal I/R injury and control groups, and macrophages M0, activated memory CD4 T cells, activated CD4 T cells, and resting NK cells were increased but do not so significantly. Meanwhile, CD8 T cells, activated NK cells, resting dendritic cell and activated mast cells were significantly reduced in intestinal I/R injury (Figure 5B).
Correlation of the coFRGs Signature and Infiltrating Immune Cells Characteristics
Spearman correlation analysis was used to reveal the correlation between coFRGs signature and immune cells. Plasma cell, CD8 T cells, resting NK cells, activated NK cells, Monocytes, Macrophages M0, Macrophages M1, activated dendritic cells were correlated with the expression profile of coFRGs from mouse, and CD8 T cells, naive T cells, resting memory CD4 T cells, activated memory CD4 T cells, activated NK cells, Monocytes, Macrophages M0, Macrophages M1, resting dendritic cells, activated dendritic cells, resting mast cells, eosinophils, neutrophils were correlated with the expression profile of coFRGs from human (Figure 5C and D).
Validation of the Hub Genes in Mouse Intestinal I/R Injury Model
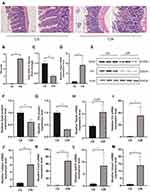
Figure 6A and B shows that the intestinal mucosal injured in the I/R model. Serum GSH is decreased after intestinal I/R (p < 0.01, Figure 6C). Exposure to the intestinal I/R injury increased the expression of Ptgs2 mRNA (p < 0.01, Figure 6D) and decreased the expression of Gpx4 (p < 0.05, Figure 6E and F) and ferritin heavy chain (Fth) (p < 0.01, Figure 6G) protein. The above results indicate that ferroptosis participates in the mechanisms underlying the intestinal I/R injury. GDF15 (p < 0.05), HMOX1 (p < 0.05), IL6 (p < 0.05), CXCL2 (p < 0.01) and TNFAIP3 (p < 0.01) were significantly increased after intestinal I/R injury (Figure 6I–M), and Hspa5 was increased but not significantly (Figure 6H).
Discussion
In the study, we discussed ferroptosis and immune-related genes after intestinal I/R injury to provide clues at the transcriptional level for subsequent studies. We extracted 61 mmu-FRGs and 294 mmu-IRGs following intestinal I/R injury in mouse, then constructed PPI network and performed functional enrichment analysis. To make the results clinically meaningful, we used transcriptome sequencing from patients with intestinal I/R injury to validate our results. We extracted 45 hsa-FRGs and 200 hsa-IRGs from human samples, resulting in 24 coFRGs and 100 coIRGs (Figure 2). Subsequently, we predicted the relationship between TFs-coFRGs and TFs-coIRGs. We also identified six hub genes from the 24 coFRGs and 100 coIRGs, predicted gene-drug pairs, and analyzed their interacting partners by Genemania database. More importantly, our findings show for the first time that immune cells are altered in the early stages of intestinal ischemia-reperfusion injury and coFRGs are closely associated with immune cells following intestinal ischemia-reperfusion injury in mouse and human.
GO results showed that mmu-FRGs were mainly involved oxidative stress, apoptotic, while mmu-IRGs were mainly related to inflammation and immunity. Meanwhile, KEGG enrichment analysis suggested that both mmu-FRGs and mmu-IRGs were enriched in IL−17 signaling pathway and TNF signaling pathway, both of which are related to immunity. The above results suggested that some mmu-FRGs and mmu-IRGs are common nodes in some pathways, and were closely linked to each other.
However, it has yet to be shown whether ferroptosis-related genes were associated with immune cells in intestinal I/R injury. Accumulating evidence has proven that ferroptosis is related to immunity, but no study has shown the interaction between ferroptosis and immunity after intestine I/R injury.10,11,23 In our study, PPI analysis showed a robust association between mmu-FRGs and mmu-IRGs, which is consistent with previous reports.10,11 In addition, Spearman correlation analysis revealed that immune cells were correlated with the expression profile of coFRGs in mice and humans.
Immune cells including macrophages M0 and activated memory CD4 T cells were increased after intestinal I/R in mouse. Meanwhile, macrophages M1 were increased and macrophages M2 were decreased in the intestinal I/R injury, which is consistent with previous research result. Inflammation is an intricate process related with an immune response. Studies showed that proinflammatory macrophage M1 polarization had played a role in I/R injury, and macrophages M2 could alleviate I/R injury.24–26 In human, immune cells including neutrophils and mast cells activated were increased significantly in intestinal I/R injury, and macrophages M0, activated memory CD4 T cells, resting NK cells were increased but do not so significantly, which was similar to mouse samples. Also, there are some differences between mouse and human. CD8 T cells, activated NK cells, resting dendritic cell and activated mast cells were decreased in human intestinal I/R injury but not mouse sample, and neutrophils were increased in human being as samples but not mouse samples. Such discrepancies in the results may be caused by the difference in collection times of the analyzed samples. The mouse samples were collected after 1 hour of ischemia followed by 4 of hours reperfusion, while the human being as sample were collected during surgery without limited ischemia and reperfusion time. This difference makes the discrepancy between experiments difficult to handle realistically. In short, intestinal I/R is an inflammation-related pathological process involving ferroptosis and immunity, but the exact mechanisms need further investigation.
In addition, we used the 6 hub genes to predict potentially effective drugs in DGIdb database, and to find protein-protein relationship in Genemania database. HSPA5, heat shock protein family A (Hsp70) member 5, may be a negative regulator of ferroptosis.27,28 Mianserin, an atypical antidepressant used in the treatment of depression, norgestrel, a synthetic progestogen, chlorquinaldol, an antimicrobial agent, and fluorouracil, a pyrimidine analogue used as an antineoplastic agent, were predicated to target HSPA5. GDF15, growth differentiation factor 15, encodes a secreted ligand of the TGF-beta (transforming growth factor-beta) superfamily of proteins which are expressed in a broad range of cell types, acts as a pleiotropic cytokine and is involved in the stress response program of cells after cellular injury.29,30 Liu et al elucidated that Gdf15 contributed to an early acting, protective injury response, modifying immune cell actions.31 Calcitriol, a synthetic physiologically-active analog of vitamin D regulating calcium in vivo by promoting absorption in the intestine, etoposide, a proliferation inhibitor, and diclofenac, a nonsteroidal benzeneacetic acid derivative with anti-inflammatory activity, target GDF15. TNFAIP3, TNF alpha induced protein 3, encoded the protein which has both ubiquitin ligase and deubiquitin enzyme activity and participates in cytokine-mediated immune and inflammatory responses. Matsuzawa’s findings illustrated that Tnfaip3 regulates survival in CD4 T cells by promoting autophagy.32 Zhang’s research implicated Tnfaip3 as a functionally important endogenous suppressor of ASK1 hyperactivation in the pathogenesis of NASH and identify it as a potential new molecular target for NASH therapy.33 From DGIdb database we found that methotrexate, an antimetabolite and antifolate agent with antineoplastic and immunosuppressant activities, and ustekinumab, a targeted antibody therapy used to manage inflammatory conditions, target TNFAIP3. HMOX1, heme oxygenase 1, is an essential enzyme in heme catabolism. Upregulation of Hmox1 promotes ferroptosis in acute lung injury, diabetic atherosclerosis and osteosarcoma cells.34–36 Sorafenib, an oral multikinase inhibitor, sunitinib, an indolinone derivative and tyrosine kinase inhibitor with potential antineoplastic activity, and aspirin, an anti-inflammation salicylate, target HMOX1. CXCL2, C-X-C motif chemokine ligand2, is part of chemokine superfamily that encodes proteins involved in immunoregulatory and inflammatory processes. Previous studies elucidating that the mast cell and macrophage chemokine CXCL2 controls the early stages of neutrophil recruitment, which is often the first recruited immune cell during tissue inflammation, results are consistent with our finding that neutrophil and mast cell activation is increased in the human intestine after intestinal I/R injury.37 We found that deferoxamine, a kind of inhibitor of ferroptosis, and alteplase, a thrombolytic agent, target CXCL2. IL6 is a pleiotropic cytokine with significant functions in regulating the immune system, and could induce ferroptosis.38–40 A number of drugs are predicted to target IL6, such as cisplatin, nelfinavir, metronidazole, rituximab, linezolid, ifosfamide, fentanyl and so on. Fentanyl, an opioid painkiller, targets IL6. Indeed, some studies have shown that opioids can alleviate ischemia-reperfusion injury.41,42
Furthermore, HSPA5 and its interacting genes were enriched in endoplasmic reticulum stress, GDF15 was mainly focus on extracellular stimulus, and HMOX1 was mainly involved in iron ion homeostasis (Figure 4D–F). TNFAIP3, CXCL2, IL6 and their interacting genes were mainly enriched in pathways associated with inflammatory and immunity, such as cytokine activity, toll-like receptor signaling pathway, cytokine receptor binding and so on (Figure 4G–I). In addition, the 6 hub genes interact with each other (Figure S9). To some extent, these genes can be used as target genes for subsequent studies. The genes-drug network gives us potential drugs to treat intestinal I/R injury. However, it should be validated in further studies.
We also validated the expression of the six hub genes by qPCR in intestinal samples from mouse intestinal I/R injury mode, and the results were generally consistent with RNA-sequencing results. In addition, ferroptosis did occur after intestinal I/R injury.
This study has several limitations. Firstly, the study mainly consisted of a database mining design without sufficient validation experimental studies, especially in clinical practice. Secondly, we did not perform validation after successful prediction of therapeutic agents, which will be taken into account in our further studies. We will conduct more experiments to confirm our findings.
In summary, this is the first study to focus on the differential ferroptosis and immune-related genes after intestinal ischemia-reperfusion injury and reveal a correlation between FRGs and immune cells. Our study shows that there are many differentially expressed genes related to ferroptosis and immunity after intestinal I/R injury and that there is a close relationship between FRG and IRG. These differentially expressed genes may be potential therapeutic targets and deserve further exploration. We look at intestinal I/R injury in terms of the combination of ferroptosis and immunity, providing a new direction for our subsequent studies.
Conclusion
In conclusion, we identified ferroptosis and immune-related genes to predict their correlations in intestinal I/R injury. We also predicted potential TF-gene networks and potential therapeutic targets (HSPA5, GDF15, TNFAIP3, HMOX1, CXCL2 and IL6), providing clues for further studies of intestinal I/R injury.
Data Sharing Statement
Data could be obtained upon request to the corresponding author.
Ethics Approval
The animal experiments were approved by the Ethics Committee of Nanfang Hospital (China, application No: NFYY-2021-54) and all procedures were carried out in compliance with National Institutes of Health guidelines for the use of experimental animals.
Acknowledgment
We thank for technical support. We thank Dr.Jianming Zeng (University of Macau), Yuzhong Peng (Macau University of Science and Technology) and all the other members of Zeng’s bioinformatics team, biotrainee, for generously sharing their experience and codes. Special thanks go to Dr. Yang for English edits.
Author Contributions
All authors have made significant contributions to the work of the report, whether in concept, study design, execution, data collection, analysis and interpretation, or in all of these areas; participating in drafting, revising, or reviewing articles; finally approving the version to be published; agreeing to the journals in which the article has been submitted; and agreeing to be responsible for all aspects of the work.
Funding
This work was supported by grants from the Key Program of National Natural Science Foundation, Beijing, China (81730058 to Ke-Xuan Liu).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Kalogeris T, Baines CP, Krenz M, Korthuis RJ. Ischemia/Reperfusion. Compr Physiol. 2016;7(1):113–170. doi:10.1002/cphy.c160006
2. Stockwell BR, Friedmann Angeli JP, Bayir H, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171(2):273–285. doi:10.1016/j.cell.2017.09.021
3. Müller T, Dewitz C, Schmitz J, et al. Necroptosis and ferroptosis are alternative cell death pathways that operate in acute kidney failure. Cell Mol Life Sci. 2017;74(19):3631–3645. doi:10.1007/s00018-017-2547-4
4. Tuo QZ, Lei P, Jackman KA, et al. Tau-mediated iron export prevents ferroptotic damage after ischemic stroke. Mol Psychiatry. 2017;22(11):1520–1530. doi:10.1038/mp.2017.171
5. Li Y, Feng D, Wang Z, et al. Ischemia-induced ACSL4 activation contributes to ferroptosis-mediated tissue injury in intestinal ischemia/reperfusion. Cell Death Differ. 2019;26(11):2284–2299. doi:10.1038/s41418-019-0299-4
6. Fang X, Wang H, Han D, et al. Ferroptosis as a target for protection against cardiomyopathy. Proc Natl Acad Sci U S A. 2019;116(7):2672–2680. doi:10.1073/pnas.1821022116
7. Deng F, Zhao BC, Yang X, et al. The gut microbiota metabolite capsiate promotes Gpx4 expression by activating TRPV1 to inhibit intestinal ischemia reperfusion-induced ferroptosis. Gut Microbes. 2021;13(1):1–21. doi:10.1080/19490976.2021.1902719
8. Li J, Cao F, Yin HL, et al. Ferroptosis: past, present and future. Cell Death Dis. 2020;11(2):88. doi:10.1038/s41419-020-2298-2
9. Jiang P, Yang F, Zou C, et al. The construction and analysis of a ferroptosis-related gene prognostic signature for pancreatic cancer. Aging. 2021;13(7):10396–10414. doi:10.18632/aging.202801
10. Proneth B, Conrad M. Ferroptosis and necroinflammation, a yet poorly explored link. Cell Death Differ. 2019;26(1):14–24. doi:10.1038/s41418-018-0173-9
11. O’Donnell VB, Aldrovandi M, Murphy RC, Krönke G. Enzymatically oxidized phospholipids assume center stage as essential regulators of innate immunity and cell death. Sci Signal. 2019;12(574):eaau2293. doi:10.1126/scisignal.aau2293
12. Yan HF, Tuo QZ, Yin QZ, Lei P. The pathological role of ferroptosis in ischemia/reperfusion-related injury. Zool Res. 2020;41(3):220–230. doi:10.24272/j.issn.2095-8137.2020.042
13. Jiang Z, Chen S, Zhang L, Shen J, Zhong M. Potentially functional microRNA-mRNA regulatory networks in intestinal ischemia-reperfusion injury: a bioinformatics analysis. J Inflamm Res. 2021;14:4817–4825. doi:10.2147/JIR.S328732
14. Zhou N, Bao J. FerrDb: a manually curated resource for regulators and markers of ferroptosis and ferroptosis-disease associations. Database. 2020;2020:baaa021. doi:10.1093/database/baaa021
15. Raivo K. pheatmap: pretty Heatmaps. R package version 1.0.12; 2019. Available from: https://CRAN.R-project.org/package=pheatmap.
16. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
17. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi:10.1089/omi.2011.0118
18. Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi:10.1093/nar/gkq537
19. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi:10.1038/nmeth.3337
20. Deng F, Hu J, Yang X, et al. Interleukin-10 expands transit-amplifying cells while depleting Lgr5+ stem cells via inhibition of Wnt and notch signaling. Biochem Biophys Res Commun. 2020;533(4):1330–1337. doi:10.1016/j.bbrc.2020.10.014
21. Zhou B, Zhang W, Yan Z, et al. MicroRNA-26b-5p targets DAPK1 to reduce intestinal ischemia/reperfusion injury via inhibition of intestinal mucosal cell apoptosis. Dig Dis Sci. 2021. doi:10.1007/s10620-021-06975-7
22. Zhang XY, Liu ZM, Wen SH, et al. Dexmedetomidine administration before, but not after, ischemia attenuates intestinal injury induced by intestinal ischemia-reperfusion in rats. Anesthesiology. 2012;116(5):1035–1046. doi:10.1097/ALN.0b013e3182503964
23. Ganz T, Nemeth E. Iron homeostasis in host defence and inflammation. Nat Rev Immunol. 2015;15(8):500–510. doi:10.1038/nri3863
24. Zhang M, Nakamura K, Kageyama S, et al. Myeloid HO-1 modulates macrophage polarization and protects against ischemia-reperfusion injury. JCI Insight. 2018;3(19):120596. doi:10.1172/jci.insight.120596
25. Fan Q, Tao R, Zhang H, et al. Dectin-1 contributes to myocardial ischemia/reperfusion injury by regulating macrophage polarization and neutrophil infiltration. Circulation. 2019;139(5):663–678. doi:10.1161/CIRCULATIONAHA.118.036044
26. Kormann R, Kavvadas P, Placier S, et al. Periostin promotes cell proliferation and macrophage polarization to drive repair after AKI. J Am Soc Nephrol. 2020;31(1):85–100. doi:10.1681/ASN.2019020113
27. Zhu S, Zhang Q, Sun X, et al. HSPA5 regulates ferroptotic cell death in cancer cells. Cancer Res. 2017;77(8):2064–2077. doi:10.1158/0008-5472.CAN-16-1979
28. Chen Y, Mi Y, Zhang X, et al. Dihydroartemisinin-induced unfolded protein response feedback attenuates ferroptosis via PERK/ATF4/HSPA5 pathway in glioma cells. J Exp Clin Cancer Res. 2019;38(1):402. doi:10.1186/s13046-019-1413-7
29. Assadi A, Zahabi A, Hart RA. GDF15, an update of the physiological and pathological roles it plays: a review. Pflugers Arch. 2020;472(11):1535–1546. doi:10.1007/s00424-020-02459-1
30. Baek SJ, Eling T. Growth differentiation factor 15 (GDF15): a survival protein with therapeutic potential in metabolic diseases. Pharmacol Ther. 2019;198:46–58. doi:10.1016/j.pharmthera.2019.02.008
31. Liu J, Kumar S, Heinzel A, et al. Renoprotective and Immunomodulatory effects of GDF15 following AKI invoked by ischemia-reperfusion injury. J Am Soc Nephrol. 2020;31(4):701–715. doi:10.1681/ASN.2019090876
32. Matsuzawa Y, Oshima S, Takahara M, et al. TNFAIP3 promotes survival of CD4 T cells by restricting MTOR and promoting autophagy. Autophagy. 2015;11(7):1052–1062. doi:10.1080/15548627.2015.1055439
33. Zhang P, Wang PX, Zhao LP, et al. The deubiquitinating enzyme TNFAIP3 mediates inactivation of hepatic ASK1 and ameliorates nonalcoholic steatohepatitis. Nat Med. 2018;24(1):84–94. doi:10.1038/nm.4453
34. Dong H, Qiang Z, Chai D, et al. Nrf2 inhibits ferroptosis and protects against acute lung injury due to intestinal ischemia reperfusion via regulating SLC7A11 and HO-1. Aging. 2020;12(13):12943–12959. doi:10.18632/aging.103378
35. Lin H, Chen X, Zhang C, et al. EF24 induces ferroptosis in osteosarcoma cells through HMOX1. Biomed Pharmacother. 2021;136:111202. doi:10.1016/j.biopha.2020.111202
36. Meng Z, Liang H, Zhao J, et al. HMOX1 upregulation promotes ferroptosis in diabetic atherosclerosis. Life Sci. 2021;284:119935. doi:10.1016/j.lfs.2021.119935
37. De Filippo K, Dudeck A, Hasenberg M, et al. Mast cell and macrophage chemokines CXCL1/CXCL2 control the early stage of neutrophil recruitment during tissue inflammation. Blood. 2013;121(24):4930–4937. doi:10.1182/blood-2013-02-486217
38. Yao X, Huang J, Zhong H, et al. Targeting interleukin-6 in inflammatory autoimmune diseases and cancers. Pharmacol Ther. 2014;141(2):125–139. doi:10.1016/j.pharmthera.2013.09.004
39. Xue H, Yuan G, Guo X, et al. A novel tumor-promoting mechanism of IL6 and the therapeutic efficacy of tocilizumab: hypoxia-induced IL6 is a potent autophagy initiator in glioblastoma via the p-STAT3-MIR155-3p-CREBRF pathway. Autophagy. 2016;12(7):1129–1152. doi:10.1080/15548627.2016.1178446
40. Bin S, Xin L, Lin Z, Jinhua Z, Rui G, Xiang Z. Targeting miR-10a-5p/IL-6R axis for reducing IL-6-induced cartilage cell ferroptosis. Exp Mol Pathol. 2021;118:104570. doi:10.1016/j.yexmp.2020.104570
41. Franco-Acevedo A, Echavarria R, Moreno-Carranza B, et al. Opioid preconditioning modulates repair responses to prevent renal ischemia-reperfusion injury. Pharmaceuticals. 2020;13(11):E387. doi:10.3390/ph13110387
42. Xiao Y, Phelp P, Wang Q, et al. Cardioprotective properties of known agents in rat ischemia-reperfusion model under clinically relevant conditions: only the NAD precursor nicotinamide riboside reduces infarct size in presence of fentanyl, midazolam and cangrelor, but not propofol. Front Cardiovasc Med. 2021;8:712478. doi:10.3389/fcvm.2021.712478
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.