Back to Journals » Drug Design, Development and Therapy » Volume 15
Inhibitory Effects of Periplocin on Lymphoma Cells: A Network Pharmacology Approach and Experimental Validation
Authors Zhao R , Han C, Dai S, Wei S, Xiang X, Wang Y, Zhao R, Zhao L, Shan B
Received 18 January 2021
Accepted for publication 13 March 2021
Published 26 March 2021 Volume 2021:15 Pages 1333—1344
DOI https://doi.org/10.2147/DDDT.S302221
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Anastasios Lymperopoulos
Riyang Zhao, Chen Han, Suli Dai, Sisi Wei, Xiaohan Xiang, Yaojie Wang, Ruinian Zhao, Lianmei Zhao, Baoen Shan
Research Centre, The Fourth Hospital of Hebei Medical University, Shijiazhuang, Hebei, 050011, People’s Republic of China
Correspondence: Baoen Shan; Lianmei Zhao
Research Centre, The Fourth Hospital of Hebei Medical University, 12 Jiankang Road, Shijiazhuang, Hebei, 050011, People’s Republic of China
Tel/Fax +86 311-86095283
Email [email protected]; [email protected]
Purpose: Lymphoma is considered to be one of the most pressing health problems worldwide owing to its high incidence and mortality. Previous studies have shown that periplocin, a naturally occurring compound, inhibits growth and induces apoptosis in several cancers. However, the effects of periplocin on lymphoma and the underlying mechanisms of action remain unclear.
Methods: The PharmMapper database was used to predict the potential targets of periplocin. The GeneCard database was used to identify lymphoma-related genes. A few intersecting genes were obtained, and the protein-protein interaction network was visualized using STRING Gene ontology analysis. Kyoto Encyclopedia of Genes and Genomes pathway analyses were performed using R project. MTS assay, flow cytometry, real-time quantitative polymerase chain reaction (qPCR), and Western blotting were used to verify whether periplocin possesses anti-lymphoma activity.
Results: A total of 216 intersecting genes were identified. Numerous cancer-related signaling pathways were visualized using Cytoscape software, with the PI3K-Akt signaling pathway being the highest-ranked pathway related to cell proliferation, apoptosis, and cell cycle progression. HuT 78 and Jurkat cell lines were used to verify the predictions. Periplocin significantly inhibited their proliferation in a dose- and time-dependent manner, but had no effect on the viability of peripheral blood lymphocytes. Flow cytometry revealed that treatment with periplocin increased the apoptotic rate and ratio of HuT 78 and Jurkat cells in the G2/M phase. CDK1 and cyclin B1 complex formation is a key gatekeeper to mitotic division in the G2/M phase. Western blot analysis revealed that periplocin significantly decreased the protein levels of CDK1 and cyclin B1; however, real-time qPCR revealed no effect on gene expression.
Conclusion: Periplocin showed anti-tumor effects in lymphoma cells through multiple targets and signaling pathways, and could be a novel therapeutic agent for the treatment of lymphoma.
Keywords: periplocin, lymphoma, structure-function analysis, apoptosis, cell cycle
Introduction
Lymphoma is a malignant tumor originating from the lymphohematopoietic system, mainly manifesting as painless lymphadenectasis, hepatosplenomegaly, and fever, with other systemic symptoms. There are two main types of lymphoma: non-Hodgkin’s lymphoma (NHL) and Hodgkin’s lymphoma (HL). In 2018, 509,590 and 79,990 new cases of NHL and HL were respectively recorded worldwide. In addition, 508,585 and 26,167 deaths from NHL and HL, respectively, occurred in the same year.1 The incidence of lymphoma is rapidly increasing, with lymphoma currently ranked as one of the ten most common malignant tumors in the US.2 In China, 88,200 new cases and 52,100 lymphoma deaths were reported in 2016. Regarding risk factors, both morbidity and mortality are better for male patients than females.3 Although encouraging remission rates have arisen with optimal treatment based on standard staging, prevention must still be pursued. Consequently, the latter approach can minimize treatment-related toxicity, which is often irreversible, as well as late-onset, such as cardiac dysfunction or second malignancy induction.4 The treatment of the two most common forms of NHL, follicular lymphoma and diffuse large B cell (DLBCL), was revolutionized by the discovery of rituximab and its advantage in chemotherapeutic regimens. However, the prognosis remains poor for one-quarter of patients with follicular lymphoma, and for one-third of patients with relapsed-refractory DLBCL who are apt for transformation to a more aggressive lymphoma and/or early relapse.5 One study reported that 20% of patients with DLBCL who achieved complete remission using standard therapy eventually experienced relapse.6 Despite considerable advances in the treatment of lymphoma, the prognosis of patients with relapsed and/or refractory disease continues to be poor.7 Moreover, cases of indolent lymphomas, typically with an extended course and benign prognosis, may transform to a more aggressive phenotype, with the prognosis of transformed cases being very poor.8 To address these burdens, a new and innovative therapeutic strategy for lymphoma is crucial.
Hundreds of compounds, from steroids to cardiotonic glycosides, can be extracted from the traditional Chinese herb, Periploca sepium Bunge. Many of these compounds display fascinating and beneficial biological effects, mostly in the treatment of autoimmune diseases.9 Relevant to this research is periplocin, a small molecule that has been shown to inhibit various cancer types and display anti-tumor properties.10–13 Despite its clinical potential, the structure-function analysis of periplocin is absent from current literature. In this report, we investigate the structure of periplocin for bioinformatic predictions of the underlying mechanisms of its effects. For the first time, we used network pharmacology approaches and bioinformatic predictions to forecast the intersecting genes and hub genes of periplocin and its influence on lymphoma. The intersecting genes were established using a protein-protein intersection (PPI) network, gene ontology (GO) enrichment, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. Functional experiments were performed to verify the effects of periplocin on cell viability, cell cycle, and the apoptosis of lymphoma cell lines. Therefore, we present a reliable and relevant method for the discovery and activity determination of a novel agent possessing anti-lymphoma activity.
Materials and Methods
Preparation and Dilution of Periplocin and Vincristine
Periplocin (C36H56O13, purity ≥ 96%) (New Drug Research and Development Center of the North China Pharmaceutical Group Corporation, Hebei, China) was purified and diluted as described in our previous studies.14–16
Vincristine (C46H56N4O10, purity ≥ 98%; (VCR)) (Solarbio Science and Technology Ltd) and periplocin were dissolved in dimethyl sulfoxide (DMSO) and diluted to the desired concentration with serum-free RPMI-1640 medium (final concentration of DMSO < 0.01%).
Prediction of Periplocin Targets and Acquisition of Lymphoma-Related Genes
The three-dimensional (3D) structure of periplocin was acquired from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). The SDF file of the 3D structure was uploaded to the PharmMapper database for potential target prediction. The GeneCard database was used to acquire lymphoma-related genes (https://www.GeneCards.org/).
Acquisition of Intersecting Genes and PPI Network Construction
Potential periplocin targets and lymphoma-related genes were uploaded to the Jvenn website (http://jvenn.toulouse.inra.fr/app/example.html) to determine the intersecting genes. These genes were then imported into Cytoscape software to construct the “lymphoma-targets-periplocin” network. The PPI network containing 216 intersecting genes was created using the STRING database (https://string-db.org/), with a high confidence level (interaction score > 0.90).
GO and KEGG Pathway Analysis
The GO and KEGG pathway enrichment analyses of the intersecting genes were carried out using the R project (version 4.0.2); this displayed the 20 top-ranked biological processes of these genes.
Cell Lines and Culture
The lymphoma cell lines, HuT 78 and Jurkat (OTWO BIOTECH Company), were used in the present study. Peripheral blood lymphocytes (PBLs) were obtained from the whole blood of healthy donors using human lymphocyte separation medium (Hao Yang Biological Manufacture Company, Tianjin, China). No procedures were performed on participant donors until written consent was obtained from all subjects or guardians. Three (3) mL each of separation medium followed by whole blood was added to a centrifuge tube for centrifugation at 500 g for 25 min. As a result, PBLs were separated into the annular opalescent lymphocyte layer. PBLs were extracted and washed with phosphate buffered saline (PBS). All cells were cultured in 5% CO2 at 37 °C in RPMI 1640 medium (Gibco, USA) supplemented with 10% (20% in PBL) fetal bovine serum (Biological Industries, Beit-Haemek, Israel), penicillin, and streptomycin (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA).
MTS Assay
The MTS assay (Promega, USA) was used to examine cell proliferation. HuT 78, Jurkat, and PBL cells were harvested and sub-cultured in 96-well plates (8000 cells/well). After incubating the cells for 24, 48, and 72 h, cell proliferation was assessed by an MTS assay at each time point according to the manufacturer’s instructions. MTS reagent (20 μL/well) was added to each well. The 96-well plates were incubated at 37 °C for 2 h in a dark room, and their absorbance at 492 nm was determined using a microplate reader. The experiment was performed in triplicate and repeated three times.
Flow Cytometry Assay
The apoptotic rate and cell cycle distribution of HuT 78 and Jurkat cells were detected using flow cytometry. The treated and untreated cells were harvested in PBS. For apoptosis detection, the cells were incubated with Annexin V-PE and 7-AAD (Pharmingen TM PE Annexin V Apoptosis Detection Kit I, BD, USA) for 15 min at room temperature in a dark room. For cell cycle detection, the cells were incubated with DNA staining solution and permeabilization solution (Cell Cycle Staining Kit, MULTI SCIENCES BIOTECH, China) for 30 min in the dark. The stained cells were then analyzed by flow cytometry (FACS Calibur, BD, USA).
RNA Extraction and qPCR
RNA extraction and real-time qPCR were performed according to previously reported protocols. Total RNA was extracted from cells using TRIzol reagent (Invitrogen, CA, USA). The GoScriptTM Reverse Transcription System (Promega, USA) was used to synthesize cDNA from 2 μg of total RNA. qPCR was performed in triplicate with a 1:4 dilution of cDNA using the SYBR Green PCR Kit (Promega, USA), with a subsequent real-time PCR system (ABI 7500). The gene-specific qPCR primers were as follows: CDK1 forward primer: 5ʹ-TAGTCTGGTCTTTCTTTGGCTG-3ʹ; CDK1 reverse primer: 5ʹ-GTTCAAAACTGGAATAAAACACCTA-3ʹ; cyclin B1 forward primer: 5ʹ-GCAGCACCTGGCTAAGAATGT-3ʹ; Cyclin B1 reverse primer: 5ʹ-GCCTTGGCTAAATCTTGAACT-3ʹ; and GAPDH forward primer: 5ʹ- GGACCTGACCTGCCGTCTAG-3ʹ; GAPDH reverse primer: 5ʹ- GTAGCCCAGGATGCCCTTGA-3ʹ.
Docking Periplocin to Cyclin B1 and CDK1
The 3D structure of periplocin was drawn using ChewDraw software (version Professional 18.0). The 3D structures/X-ray crystal structures of cyclin B1 and CDK1 were obtained from the Protein Data Bank (www.rcsb.org). Docking of periplocin to cyclin B1 and CDK1 was performed using AutoDock software (version 4.2). The images of docking periplocin to cyclin B1 and CDK1 were presented using PyMol software (version 2.2.0).
Western Blot
After RIPA with PMSF (Solarbio, China) was used to split cells, proteins were separated by 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred electrophoretically onto a PVDF membrane (Millipore, Billerica, MA, USA). The mixture was incubated in 5% skim milk at room temperature (25 °C) for 1 h. The membrane was incubated with antibodies against CDK1, cyclin B1 (Proteintech, China), and GAPDH (Bioworld, USA) at 4 °C for 12 h. Fluorochrome-labeled secondary anti-rabbit IgG was then used to incubate the membranes at room temperature (25 °C) for 1 h. The membranes were visualized using the Odyssey Infrared Imaging System (LI-COR, USA). GAPDH served as the loading control. Western blot analysis was independently performed three times.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism (version 5.0) and SPSS (IBM, version 22.0). Data are presented as mean ± standard deviation (SD). One-way ANOVA was used to examine significant differences in the experimental data between the groups. P values indicated in each figure, including *p < 0.05, **p < 0.01, ***p < 0.001, indicate significance. As previously mentioned, all experiments were independently repeated thrice.
Results
Determination of Lymphoma-Related Genes That Might Be Regulated by Periplocin
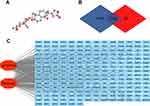
The 3D structure of periplocin (Figure 1A) was obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). After uploading the structure to the PharmMapper database (http://lilab-ecust.cn/pharmmapper/), 299 potential binding genes were acquired from the platform (Table S1). A total of 7592 lymphoma-related genes were found in the GeneCard database (https://www.GeneCards.org/) (Table S2). There were 216 intersecting genes through which periplocin may regulate the progression of lymphoma (Figure 1B) (Table S3).17 The potential intersection network is depicted in Figure 1C.
Construction of a PPI Network
To further investigate the relationship between periplocin and lymphoma, the STRING database (https://string-db.org/) was used to construct the PPI network. After uploading the 216 intersecting genes to the STRING database, 216 nodes and 542 intersecting relationships were identified. The resulting PPI network is shown in Figure 2A. After importing the network into the Cytoscape software, 10 hub genes (ranked by the MCC method) with potentially crucial pathway roles were obtained: MAPK1, LCK, PTPN11, RHOA, SRC, IL2, JAK3, EGFR, AKT1, and JAK2 (Figure 2B) (Table S4).
 |
Figure 2 The construction of a PPI network. (A) The PPI network of the 216 intersecting genes from Figure 1B. (B) The hub genes of the PPI network. The darker the red, the higher the degree of connection. |
GO and KEGG Pathway Analyses
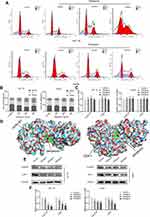
The GO and KEGG pathway analyses were performed by the R Project (version 4.0.2) to uncover the underlying mechanisms of the intersecting genes through which periplocin may regulate lymphoma. As shown in Figure 3A, the biological process enrichment analysis revealed that periplocin may regulate lymphoma through neutrophil-mediated immunity, neutrophil activation, and T cell activation. Furthermore, cellular component enrichment showed that periplocin may impact the vesicle lumen, cytoplasmic vesicle lumen, secretory granule lumen, and membrane rafts (Figure 3B). Molecular function enrichment suggested that periplocin may act on protein serine/threonine kinase activity, protein tyrosine kinase activity, endopeptidase activity, and carboxylic acid binding (Figure 3C).
The results of KEGG analysis revealed that the PI3K-Akt, Ras, and MAPK signaling pathways were the top three highest-ranked potential pathways (Figure 3D), with 28, 24, and 24 intersecting genes enriched, respectively. The KEGG network analysis is shown in Figure 3E. The intersecting genes enriched in the PI3K-Akt pathway (Figure 3F),18 such as eNOS and ERK, are related to cell proliferation, angiogenesis, and DNA repair, while PDK1, Akt, GSK3, and CDKs are associated with cell progression. In the MAPK pathway, ERK is correlated to proliferation and differentiation, while AKT, JNK, and p38 are involved in proliferation, differentiation, inflammation, and apoptosis (data not shown). These results suggest that periplocin may influence cell proliferation, apoptosis, and cell cycle progression in lymphoma. To confirm these findings, additional experiments were performed as described below.
Effects of Periplocin on the Lymphoma Cell Lines and PBLs
The MTS assay was used to verify the effect of periplocin on lymphoma proliferation. HuT 78 or Jurkat cells were cultured in 96-well plates with different concentrations of periplocin, with 400 ng/mL VCR as the positive control. As shown in Figure 4A, the rates of inhibition of 50, 100, 200, and 400 ng/mL periplocin and VCR in HuT 78 cells were 13.21 ± 1.63%, 26.10 ± 2.23%, 35.73 ± 1.53%, 37.93 ± 0.98%, and 10.04 ± 0.58% over 24 h, respectively. The rates of inhibition of the same concentrations of periplocin and VCR in Jurkat cells were 13.45 ± 2.60%, 23.30 ± 1.79%, 32.78 ± 0.59%, 41.66 ± 1.97%, and 14.42 ± 0.43%, respectively. At 48 hours, the rates of inhibition of 50, 100, 200, 400 ng/mL periplocin and VCR in HuT 78 cells were 23.00 ± 1.26%, 41.56 ± 0.82%, 43.88 ± 4.14%, 53.15 ± 1.76%, and 25.38 ± 3.08%, respectively. The corresponding inhibitory rates in Jurkat cells were 23.38 ± 1.66%, 25.49 ± 2.65%, 44.26 ± 2.40%, 53.09 ± 3.72%, and 26.75 ± 1.60%, respectively. At 72 hours, the inhibitory rates of 25, 50, 100, 200, 400 ng/mL periplocin and VCR in HuT 78 cells were 12.66 ± 2.48%, 25.08 ± 2.98%, 44.14 ± 2.66%, 47.00 ± 1.78%, 56.13 ± 2.30%, and 33.17 ± 0.85%, respectively. The corresponding rates of inhibition of periplocin and VCR in Jurkat cells at that time were 15.26 ± 3.55%, 33.99 ± 3.31%, 34.22 ± 3.87%, 46.67 ± 2.78%, 57.19 ± 2.72%, and 36.00 ± 2.78%, respectively. The IC50 values of periplocin in HuT 78 and Jurkat cells were 484.94 ± 24.67 and 541.68 ± 58.47 ng/mL, respectively. Such finding revealed that periplocin significantly inhibited the proliferation of lymphoma cell lines.
To explore whether periplocin had a similar effect on normal cells, PBLs from healthy donors were cultured in RPMI 1640 supplemented with 20% FBS and different concentrations of periplocin. The results showed that periplocin had no effect on the proliferation of PBLs, even after 72 h of experimentation (Figure 4B).
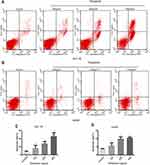
Effect of Periplocin on the Apoptosis of the Lymphoma Cell Lines
As the 100, 200, and 400 ng/mL periplocin concentrations had the most significant inhibitory effect on the proliferation of lymphoma cell lines, they were selected for flow cytometry experiments. This study aimed to determine the promotional role of periplocin in the apoptosis of lymphoma cell lines (Figure 5A and B). The apoptotic rates of HuT 78 cells treated with 100, 200, and 400 ng/mL periplocin were 16.43% ± 7.08%, 27.92% ± 5.15%, and 45.90% ± 8.69% (Figure 5C), while the corresponding apoptotic rates of Jurkat cells were 5.77 ± 1.83%, 10.11 ± 1.12%, and 10.61 ± 0.50%, respectively (Figure 5D).
Effects of Periplocin in the G2/M Phase and on CDK1 and Cyclin D1
As shown in Figure 3F,18 periplocin was predicted to influence the activity of the CDK proteins. Therefore, we hypothesized that periplocin may arrest the cell cycle of HuT 78 and Jurkat cells, and proceeded to carry out flow cytometry to verify this prediction. As shown in Figure 6A, the cell cycles of HuT 78 and Jurkat cells were arrested in the G2/M phase. The percentages of HuT 78 cells treated with 100, 200, and 400 ng/mL periplocin arrested in the G2/M phase were 23.38 ± 1.71%, 28.36 ± 5.13%, and 41.15 ± 8.48%, respectively. The corresponding percentages of Jurkat cells were 16.41% ± 1.79%, 25.39% ± 2.70%, and 24.28 ± 1.63%, respectively. All percentages were significantly higher than those of the negative control, at 15.13 ± 0.53% and 15.48 ± 0.65% (Figure 6B).
CDK1, a member of the CDK family, is regulated by cyclin B1, which forms a complex that enables G2/M phase conversion. The above results suggest that periplocin can block the cell cycle of HuT 78 and Jurkat cells at the G2/M stage and is predicted to target CDKs. Owing to this finding, researchers must aim to explore whether periplocin affects the expression of CDK1 and cyclin B1. The results obtained from qPCR suggested that periplocin had no effect on the mRNA expression of CDK1 and cyclin B1 (Figure 6C). To verify whether periplocin could directly affect cyclin B1 and CDK1, molecular docking was used in the initial survey. As shown in Figure 6D, periplocin occupied the pockets of cyclin B1 and CDK1 like a “wedge”. To confirm the inhibitory action of periplocin, Western blotting was performed. The results (Figure 6E) verified that the compound could suppress the expression of CDK1 and cyclin B1 at the protein level, with protein levels of cyclin B1 and CDK1 in HuT 78 cells treated with 100, 200, and 400 ng/mL periplocin being 85.82 ± 8.18%, 36.02 ± 6.64%, 27.13 ± 4.73%, and 72.29 ± 5.21%, 47.73 ± 5.94%, and 33.30 ± 6.40%, respectively, compared to the negative control. Moreover, the corresponding protein levels of cyclin B1 and CDK1 in Jurkat cells at the same periplocin concentrations were 70.68 ± 5.24%, 63.86 ± 3.05%, 56.34 ± 6.98%, and 82.14 ± 4.73%, 71.14 ± 6.66%, and 44.81 ± 4.85%, respectively, compared to the negative control (Figure 6F). All cyclin B1 and CDK1 protein levels treated with periplocin were significantly lower than those in the negative control (p < 0.05).
Discussion
Previously, we postulated that periplocin may indeed induce apoptosis in GC cells via the activation of the ERK1/2-EGR1 pathway.10 Furthermore, the combination of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and periplocin had a greater inhibitory effect on both gastric cancer and esophageal squamous cell carcinoma cell viability than periplocin or TRAIL alone, both in vitro and in vivo.15,16 Periplocin also inhibits the growth of colon cancer cells and downregulates the expression of c-myc, both in vitro and in vivo, via beta-catenin/TCF signaling.14
The current theory suggests that periplocin promotes the induction of cell apoptosis, inhibits proliferation and migration, and suppresses tumor development by inducing apoptosis and autophagy.10–12,14–16,19,20 However, there was no existing structure-function analysis of periplocin’s actions prior to our study. In this study, several biological information analysis databases and software were utilized to explore the possible targets of periplocin. We performed bioinformatic analysis to obtain 216 intersecting genes, which were uploaded into the STRING database to construct a PPI network. This was then imported into the Cytoscape software for 10 hub genes: MAPK1, LCK, PTPN11, RHOA, SRC, IL2, JAK3, EGFR, AKT1, and JAK2. The hub genes are related to the T cell receptor signaling pathway, B cell receptor signaling pathway, autophagy, natural killer cell-mediated cytotoxicity, human T-cell leukemia virus 1 infection, apoptosis, chronic myeloid leukemia, acute myeloid leukemia, viral carcinogenesis, cellular senescence, and necroptosis. This study utilized the R project to conduct GO and KEGG analyses to determine whether periplocin could regulate the proliferation, apoptosis, and cell cycle of lymphoma cells, and perhaps influence various cancer-related signaling pathways. The identified hub genes in turn could be endemic to the three highest-ranked pathways (PI3K-Akt, Ras, and MAPK signaling pathways). In vitro experiments showed that periplocin inhibited proliferation, promoted apoptosis, and arrested the cell cycle in the G2/M phase of lymphoma cell lines by downregulating the CDK1 and cyclin B1 proteins.
Periplocins are currently believed to inhibit various cancers via multiple ways. TRAIL-resistant HCC cells can be sensitized to TRAIL through periplocin treatment via the induction of DR4 and FADD expression and caspase 3, 8, and 9 activation.19 Lohbergera et al reported that periplocin led to growth suppression and apoptosis promotion by inducing the expression of death receptors and breaks of DNA double strands in a liposarcoma cell line.12 The authors also reported that the cell cycle of human myxofibrosarcoma cells was arrested by periplocin in the G2/M phase via the ERK/p38/JNK pathway.11 Meanwhile, the repression of PC3 (prostate adenocarcinoma) and U937 (monocytic leukemia) cell growth by periplocin is thought to occur mainly by activation of caspase-dependent apoptotic pathways and the induction of cell cycle impairment in the G2/M phase without extensive cell death, respectively.21 Periplocin can also inhibit the growth of lung cancer in vitro and in vivo by downregulating proteins, such as ATP5A1, EIF5A, ALDH1, and PSMB6,20 suppressing the Akt/Erk signaling pathway, and blocking the cell cycle in the G0/G1 phase.13
In this study, we verified whether periplocin can arrest the cell cycle in the G2/M phase. However, the mechanism of this phenomenon is not straightforward. Firstly, different cell lines may be affected by periplocin in unique ways. In addition, periplocin itself may be a multi-talented cell cycle suppressor that inhibits multiple CDKs and cyclins; the deficiency of poor targeting is a common problem among natural compounds. Inarguably, further study of the mechanism whereby periplocin arrests different cell lines in different phases is warranted.
Besides cancer cell inhibition, treatment with periplocin has been shown to decrease cell viability and the expression of cytokines, and promote apoptosis of TNF-α-induced rheumatoid arthritis fibroblasts (eg synoviocytes) through inhibition of the NF-κB signaling pathway.22 Periplocin may also promote the proliferation of microvascular endothelial cells in mice and decrease LDH release simultaneously.23 It may also boost proliferation, migration, and stimulate collagen production in fibroblast L929 cells, which is dependent on Src/ERK and PI3K/Akt pathway activation, a process mediated by Na/K-ATPase, and thus promote wound healing.24
The main obstacles to the clinical use of periplocin, which have gone unaccounted for, include cardiac toxicity reactions, such as ventricular premature beat, slow heart rate, atrioventricular block, and even death.9 Further studies need to be performed to understand the mechanisms of periplocin’s protective and toxic effects, to promote advances in treatment and enhance biological knowledge.
Conclusions
Our study demonstrated the anti-lymphoma effects of periplocin, elicited through multiple targets and signaling pathways. Periplocin suppressed proliferation, induced apoptosis, and arrested cell cycle progression in the G2/M phase by decreasing the levels of cell cycle-related proteins (CDK1 and cyclin B1) in lymphoma cells; however, this compound had no effect on the viability of PBLs.
Acknowledgment
We appreciate the language assistance provided by Grace Curran (Knight Cancer Institute, Oregon Health & Science University).
Funding
This study was supported by the National Natural Science Foundation of China (Grant numbers 81973520 and 81673642).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Teras LR, DeSantis CE, Cerhan JR, Morton LM, Jemal A, Flowers CR. 2016 US lymphoid malignancy statistics by World Health Organization subtypes. CA Cancer J Clin. 2016;66(6):443–459. doi:10.3322/caac.21357
3. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
4. Connors JM, Cozen W, Steidl C, et al. Hodgkin lymphoma. Nat Rev Dis Primers. 2020;6(1):61.
5. Heward JA, Kumar EA, Korfi K, Okosun J, Fitzgibbon J. Precision medicine and lymphoma. Curr Opin Hematol. 2018;25(4):329–334. doi:10.1097/MOH.0000000000000437
6. Armitage JO, Gascoyne RD, Lunning MA, Cavalli F. Non-Hodgkin lymphoma. Lancet. 2017;390(10091):298–310. doi:10.1016/S0140-6736(16)32407-2
7. Mugnaini EN, Ghosh N. Lymphoma. Prim Care. 2016;43(4):661–675. doi:10.1016/j.pop.2016.07.012
8. Godfrey J, Leukam MJ, Smith SM. An update in treating transformed lymphoma. Best Pract Res Clin Haematol. 2018;31(3):251–261. doi:10.1016/j.beha.2018.07.008
9. Li A, Guo X, Xie J, et al. Validation of biomarkers in cardiotoxicity induced by Periplocin on neonatal rat cardiomyocytes using UPLC-Q-TOF/MS combined with a support vector machine. J Pharm Biomed Anal. 2016;123(10):179–185. doi:10.1016/j.jpba.2016.02.014
10. Li L, Zhao LM, Dai SL, et al. Periplocin extracted from cortex periplocae induced apoptosis of gastric cancer cells via the ERK1/2-EGR1 pathway. Cell Physiol Biochem. 2016;38(5):1939–1951. doi:10.1159/000445555
11. Lohberger B, Bernhart E, Stuendl N, et al. Periplocin mediates TRAIL-induced apoptosis and cell cycle arrest in human myxofibrosarcoma cells via the ERK/p38/JNK pathway. Phytomedicine. 2020;76:153262. doi:10.1016/j.phymed.2020.153262
12. Lohberger B, Wagner S, Wohlmuther J, et al. Periplocin, the most anti-proliferative constituent of Periploca sepium, specifically kills liposarcoma cells by death receptor mediated apoptosis. Phytomedicine. 2018;51(1):162–170. doi:10.1016/j.phymed.2018.10.008
13. Lu ZJ, Zhou Y, Song Q, et al. Periplocin inhibits growth of lung cancer in vitro and in vivo by blocking AKT/ERK signaling pathways. Cell Physiol Biochem. 2010;26(4–5):609–618. doi:10.1159/000322328
14. Zhao L, Shan B, Du Y, Wang M, Liu L, Ren F. Periplocin from Cortex periplocae inhibits cell growth and down-regulates survivin and c-myc expression in colon cancer in vitro and in vivo via beta-catenin/TCF signaling. Oncol Rep. 2010;24(2):375–383.
15. Zhao LM, Li L, Huang Y, et al. Antitumor Effect of Periplocin in TRAIL-Resistant gastric cancer cells via upregulation of death receptor through activating ERK1/2-EGR1 pathway. Mol Carcinog. 2019;58(6):1033–1045. doi:10.1002/mc.22991
16. Han L, Dai S, Li Z, et al. Combination of the natural compound Periplocin and TRAIL induce esophageal squamous cell carcinoma apoptosis in vitro and in vivo: implication in anticancer therapy. J Exp Clin Cancer Res. 2019;38(1):501. doi:10.1186/s13046-019-1498-z
17. Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. jvenn: an interactive Venn diagram viewer. BMC Bioinform. 2014;15(1):293. doi:10.1186/1471-2105-15-293
18. Kanehisa M, Sato Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020;29(1):28–35. doi:10.1002/pro.3711
19. Cheng CF, Lu IH, Tseng HW, et al. Antitumor effect of periplocin in TRAIL-resistant human hepatocellular carcinoma cells through downregulation of IAPs. Evid Based Complement Alternat Med. 2013;2013:958025. doi:10.1155/2013/958025
20. Lu Z, Song Q, Yang J, et al. Comparative proteomic analysis of anti-cancer mechanism by periplocin treatment in lung cancer cells. Cell Physiol Biochem. 2014;33(3):859–868. doi:10.1159/000358658
21. Bloise E, Braca A, De Tommasi N, Belisario MA. Pro-apoptotic and cytostatic activity of naturally occurring cardenolides. Cancer Chemother Pharmacol. 2009;64(4):793–802. doi:10.1007/s00280-009-0929-5
22. Zhang X, Nan H, Guo J, Yang S, Liu J. Periplocin induces apoptosis and inhibits inflammation in rheumatoid arthritis fibroblast-like synoviocytes via nuclear factor kappa B pathway. IUBMB Life. 2020;72(9):1951–1959. doi:10.1002/iub.2328
23. Wang XY, Gao XM, Liu H, et al. Gene expression profiling of the proliferative effect of periplocin on mouse cardiac microvascular endothelial cells. Chin J Integr Med. 2010;16(1):33–40. doi:10.1007/s11655-010-0033-z
24. Chen L, Jiang P, Li J, et al. Periplocin promotes wound healing through the activation of Src/ERK and PI3K/Akt pathways mediated by Na/K-ATPase. Phytomedicine. 2019;57:72–83. doi:10.1016/j.phymed.2018.12.015
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.