Back to Journals » OncoTargets and Therapy » Volume 13
Inhibition of VEGFA Increases the Sensitivity of Ovarian Cancer Cells to Chemotherapy by Suppressing VEGFA-Mediated Autophagy
Authors Li X, Hu Z, Shi H , Wang C, Lei J, Cheng Y
Received 19 March 2020
Accepted for publication 1 July 2020
Published 18 August 2020 Volume 2020:13 Pages 8161—8171
DOI https://doi.org/10.2147/OTT.S250392
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Xia Li,1 Zhenhua Hu,2 Huirong Shi,2 Cong Wang,1 Jia Lei,1 Yan Cheng1
1Gynecological Oncology Radiotherapy Ward, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan Province, People’s Republic of China; 2Department of Gynecology and Obstetrics, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan Province, People’s Republic of China
Correspondence: Yan Cheng
The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan Province, People’s Republic of China
Tel/ Fax +86 371 66279032
Email [email protected]
Background: Ovarian cancer (OvCa) is the leading cause of death of gynecological malignancies worldwide. Vascular endothelial growth factor A (VEGFA), the most potent angiogenic factor, is responsible for tumor growth and angiogenesis, but its role in OvCa chemotherapy resistance remains unclear.
Methods: RT-PCR and Western blot were used to detect VEGFA expression in tumor cells and normal ovarian surface epithelial cells. Gene Ontology (GO) enrichment analysis was used to analyze GO terms correlated with VEGFA. In in vitro experiments, we knockdown VEGFA in tumor cells and detected the tumor cell viability and apoptosis after chemotherapy drug treatment by MTT assay and flow cytometry. Western blot was used to detect autophagy and apoptosis related proteins.
Results: We proved that VEGFA was highly expressed in tumor cells comparted with normal ovarian surface epithelial cells, and enriched GO analysis of VEGFA showed that VEGFA was involved in anti-apoptotic process. Further in vitro experiments confirmed that expression of VEGFA was correlated with chemotherapy resistance and this effect was mediated by autophagy. Meanwhile tumor cells treated with chemotherapy drugs also promoted the expression of VEGFA. Knockdown VEGFA inhibited autophagy of tumor cells and thus potents the killing efficiency in DDP resistant tumor cells and this effect could be reversed by the addition of recombinant VEGFA.
Conclusion: Taken together, our study demonstrates that VEGFA is involved in anti-apoptosis of tumor cells to chemotherapy, killing partly through autophagy, indicating that VEGFA may serve as a potential target to improve chemotherapy treatment.
Keywords: ovarian cancer, VEGFA, chemotherapy, autophagy
Introduction
Ovarian cancer is one of the most common cancers in women. It is also one of the most lethal gynecological cancers due to frequent late-stage diagnosis and disease heterogeneity.1 The standard treatment for OvCa includes surgery followed by chemotherapy. Initially, OvCa is chemotherapy-sensitive, and up to 80% of patients achieve remission following primary surgery and adjuvant platinum-based chemotherapy. However, approximately 75% of patients who respond to such therapy ultimately experience a relapse from recurrent and drug-resistance.2–5 Additional chemotherapy using platinum-and paclitaxel-based therapies are often required, but their effectiveness becomes limited in more aggressive forms of OvCa.6 Interventions that can overcome drug resistance and improve OvCa therapy are urgently required.
VEGFA is known to mediate vasculogenesis and angiogenesis by regulating the activity of endothelial cells.7 Accumulating evidence suggests that VEGFA is highly expressed in a range of human cancers, including liver, ovary, kidney, and colon cancers and is associated with tumor progression and poor prognosis.8–11 In addition to regulating angiogenesis, VEGFA also promotes tumor growth, metastasis, and survival directly.7 In OvCa, VEGFA overexpression is associated with poor survival and has been proposed as a biomarker for subsets of advanced ovarian tumors.12 Various therapies against VEGFA have been used for anti-cancer treatment.4,13 However, the role of VEGFA in chemotherapy resistance is still not clear.
A variety of chemotherapy resistance mechanisms have been reported in tumors. They include increased repair of DNA damage and decreased intracellular drug accumulation.14 Recent studies have also proved that autophagy can prevent the killing effect of chemo- or radio-therapy on tumors.15,16 Autophagy, the process of self-digestion of large macromolecules, including organelles in the cytoplasm of cells, plays a vital role in cell growth and function. The underlying mechanisms through which autophagy contributes to chemotherapy resistance is not very clear. Furthermore, the correlation between VEGFA and autophagy in ovarian cancer is not well understood.
In this study, we found that VEGFA promoted chemotherapy resistance of OvCa cells through autophagy. We found a strong correlation among VEGFA expression, autophagy induction, and OvCa cancer cell chemoresistance. These results highlight the importance of VEGFA in chemotherapy resistance and indicate that VEGFA is potential therapeutic target in OvCa.
Materials and Methods
Cells and Reagents
HOSEpiC (Catalog No.7310) was obtained from ScienCell Research Laboratories (San Diego, CA). OAW42 cells were purchased from the Cell Banks in Cancer Institute and Cancer Hospital, Chinese Academy of Medical Sciences (Beijing, China). The human ovarian adenocarcinoma cell lines, OVCAR3, CAOV3, and SKOV3, were provided by the China Center for Type Culture Collection (Wuhan, China). OvCa cell lines (A2780 and JHOS4) and cisplatin (DDP)-resistant OvCa lines (SKOV3/DDP and CAOV3/DDP) were purchased from the Cell Biology of the Chinese Academy of Sciences (Shanghai, China). DDP, Adriamycin (ADM), and Paclitaxel (PTX) were purchased from Sigma Corporation. Bafilomycin A1 (ab120497) and Recombinant VEGFA (ab117230) were purchased from Abcam Corporation. 3- (4, 5- dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide [MTT, (M2128)] was obtained from the Sigma Corporation and used as a measure of cell viability. B27 (17,504–044), epidermal growth factor (RP-8661), and bFGF (RP-8628) were purchased from Invitrogen. Heparin (9041–08-1) was purchased from Sigma-Aldrich.
Antibodies
Anti-VEGFA (ab1316), anti-caspase-3 (ab179517) antibodies, and anti-cleaved caspase-3 (ab2302) were purchased from Abcam (USA). Anti-p62 antibodies were purchased from Santacruz (CA). Anti-LC3B/MAP1LC3B (NB100-2220) and anti-Beclin1/ATG6 (NB500-249) antibodies were purchased from Novus Corporation. Anti-β-actin antibodies were purchased from Santacruz (CA). Annexin V-FITC Apoptosis Detection Kit (#4830-01-k) were purchased from R&D Systems (USA).
Bioinformatics Analyses
The Cancer Genome Atlas (TCGA) level 3 OvCa mRNA sequencing data were downloaded from UCSC Xena (http://xena.ucsc.edu/). The expression of VEGFA in tumor and normal tissue, including normal tissue from the Genotype-Tissue Expression (GTEx) database were analyzed with an online website Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/).17 For Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses, the genes correlated with VEGFA were first calculated using R and the top 200 genes selected to perform GO and KEGG analysis on DAVID website (https://david.ncifcrf.gov/). The RNAs-seq and microarray data of tumor cell lines were downloaded from Cancer Cell Line Encyclopedia (CCLE, https://portals.broadinstitute.org/ccle/about).18 The pheatmap R package was used to perform heatmap analysis.
Short Hairpin RNA (shRNA) Transfection
shRNA targeting sequence were purchased from Gene Pharma Company (Shanghai, China), the shVEGFA sequence were forward: 5′-ccggGCAGATTATGCGGATCAAACCTTCAAGAGAGGTTTGATCCGCATAATCTGCTTTTTTGGTACC-3, reverse: 3′-CGTCTAATACGCCTAGTTTGGAAGTTCTCTCCAAACTAGGCGTATTAGACGAAAAAACCATGGttaa-5′, NCshRNA: forward: 5′-ccggGTTCTCCGAACGTGTCACGTCAAGAGATTACGTGACACGTTCGGAGAATTTTTTGGTACC-3′, reverse: 3′-CAAGAGGCTTGCACAGTGCAGTTCTCTAATGCACTGTGCAAGCCTCTTAAAAAACCATGGttaa-5′. The shRNA was inserted into the pGV248-hu6-GFP+ Puro vector plasmid. For shRNA transfection, the tumor cells were seeded into the 6 wells for 24h, and 500 ul of the pGV248-hu6-shVEGFA or pGV248-hu6-shNC lentiviruses added before incubation for 48h. The VEGFA expression was confirmed by Western blotting.
Real-Time PCR (RT-PCR) Analysis
RT-PCR was used to determine VEGFA mRNA levels, as previously described.19 Briefly, the OvCa cell lines A2780, OAW42, SKOV3, JH0S4, OVCAR3 and CAOV3, and non-OvCa HOSEpiCs were seeded into 24-well plates and allowed to adhere for 8 h. Total RNA was extracted using RNA pure kits (Bioteke, Beijing, China) and cDNA generated using MLV- reverse transcriptase (Invitrogen Inc, Carlsbad, USA) at 50 °C for 30 min. RT-PCR amplifications were performed using the Applied Biosystems 7500 RT-PCR System (ABI, Foster City, USA). The reaction conditions were as follows: 95 °C for 15 min, 35 cycles of denaturation at 94 °C for 1 min, annealing at 53.6 °C for 30 s, and extension at 72 °C for 30 s, followed by a final extension at 72 °C for 5 mins. The PCR primers included: VEGFA: 5ʹ-CTCACCAAGGCCAGCACATAGG-3ʹ (forward) and 5ʹ-ATCTGGTTCCGAAAACCCTGAG-3ʹ (reverse). Β-actin: 5ʹ -GAGAAGATGACCCAGATCATGT—3ʹ (forward) and 5ʹ-ACTCCATGCCCAGGAAGGAAGG-3ʹ (reverse).
Cell Viability Assays
Cell viability was measured using MTT assays as previously described.20 The IC50 values of cytotoxicity were assessed using Graphpad 5.0 software. Briefly, OvCa cells were seeded into 96-well plates for 48 hours. Cells were then treated with various DDP concentrations (1–50 uM) for 24, 48, or 72 hours. The MTT reagent (20 µL per well, 5 mg/mL) was added to each well and incubated for 40 minutes. The media was then removed, and 150µL of DMSO added in each well to dissolve the residual MTT reagent. Absorbance values were measured at 490 nm to determine cell viability.
Sphere Formation Assay
SKOVE, SKOV3/DDP, CAOV3, and CAOV3/DDP cells were harvested and re-suspended into a single cell suspension. After that, cells were filtered with a 40μm sieve. Cells were plated in a ultra-low attachment at a density of 2x103 mL with DDP (5uM), serum-free DMEM/F12 (Gibco) medium supplemented with 2% B27, 20 ng/mL epidermal growth factor,20 ng/mL basic fibroblast growth factor, and 4 μg/mL heparin for 7 days. The spheres were photographed using an inverted fluorescence microscope.
Western Blot Analysis
The OvCa cells were seeded into 48-well plates for 8h and treated with the chemotherapy drugs, DDP, PTX, and ADM (50, 40, and 20 µM). The cells were incubated for 48 h after which they were washed 3 times in PBS. The lysis buffer containing with protease and phosphatase inhibitors were added into the wells and incubated on ice for 5 min. A total of 15 μg (15 μL volume) proteins were added into the gel and separated by 10% SDS-PAGE electrophoresis before transferring to PVDF membranes. Membranes were blocked in 5% non-fat dry milk in TBST and probed with primary antibodies at 4 °C for 12–18h. The membranes were then washed with TBST and labeled with the appropriate HRP-conjugated secondary antibodies at room temperature for 1 h. The membranes were exposed to highly sensitive ECL luminescent liquid and imaged using the Bio-Rad ChemiDoc™ XRS+ System.
Apoptosis Assays
SKOV3/DDP or shVEGFA SKOV3/DDP cells (6×105 cells/well) were seeded into six-well plates for 48 hours under the drug treatment. Cells (adherent and non-adherent) were collected and washed in ice-cold PBS. They were then stained with Annexin V-FITC, followed by PI staining for 15 minutes in the dark using the Annexin V-FITC/PI Apoptosis Detection Kit. Apoptotic cells were detected using FACs flow cytometry (BD, San Diego, CA, USA, FACSCanto II).
Statistical Analysis
Statistical Package for Social Science version 17.0 for Windows (SPSS, Chicago, IL, USA) and GraphPad Prism was used for all statistical analyses. Data were presented as the Mean ± Standard Deviation (SD). Differences between and among test groups were compared using t-test and one-way ANOVA, respectively. Independent groups were compared using the two-tailed Student’s t-test. A p-value of p<0.05 (P* <0.05, P** <0.01) was deemed statistically significant.
Results
VEGFA is Highly Expressed in OvCa Cells
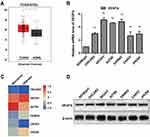
VEGFA mRNA expression levels in OvCa tumor and normal tissue were analyzed to explore the expression of VEGFA in OvCa. RNA-sequencing data from TCGA and GTEx revealed that higher expression of VEGFA in tumor tissue than normal tissue (Figure 1A). From the RT-PCR analysis results, all OvCa cells displayed increased expression of VEGFA at the mRNA level (≥ 2.5 fold increase in OVCAR3, JHOS4 and CAOV3 cells, and ≥ 4 fold increase in SKOV3, A2780, and OAW42 cells, p<0.05) compared to non-OvCa HOSEpiC cells (Figure 1B). Additionally, SKOV3 and CAOV3 cells had the highest and lowest expression of VEGFA, respectively. These results were confirmed by tumor cell line RNA-sequencing and microarray data from the CCLE database (Figure 1C). To further confirm the expression of VEGFA, Western blot analysis was used to detect VEGFA protein expression in all OvCa cell lines and HOSEpiCs cells (Figure 1D). Consistent with the above results, VEGFA was highly expressed in tumor cells.
High Expression of VEGFA Increased Chemotherapy Resistance of OvCa Cells
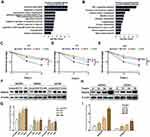
To investigate the function of VEGFA on tumor progression, the top 200 genes that positively correlated with VEGFA in OvCa from the TCGA database were selected. The GO enrichment analysis of these genes revealed some known biological functions of VEGFA, including response to hypoxia, negative regulation of the apoptotic process, and angiogenesis (Figure 2A). The KEGG analysis showed that VEGFA was mainly involved in HIF-1 signaling pathways (Figure 2B). This suggests that VEGFA is responsible for tumor growth. Chemotherapy resistance is the major cause of treatment failure; thus, we selected OvCa cells with different VEGFA expression (SKOV3, A2780, CAOV3), and treated them with the chemotherapy drugs DDP (20µM), PTX (40µM), and ADM (20µM) for 48 hours. The expression level of VEGFA was positively correlated with chemotherapy resistance in terms of tumor cells with higher expression of VEGFA was more resistant to DDP (Figure 2C–E). Moreover, we observed from the densitometry analysis of Western blots that DDP, PTX, and ADM treatment resulted in at least a 2-fold upregulation of VEGFA expression in tumor cells (Figure 2F and G). These effects were further confirmed through the assessment of the time-dependent increase of VEGFA levels in SKOV3 and CAOV3 cells treated with DDP (20µM) for 12, 24, and 48 hours. The VEGFA expression increased with time in both cell types (Figure 2H and I). These results demonstrate that VEGFA expression contributes to chemotherapy resistance and can be induced by chemotherapeutic drugs. This could be reflecting the adaptive stress responses by which tumor cells protect themselves from being killed by the chemotherapy drug.
VEGFA Knockdown Promotes DDP to Kill Tumor Cells
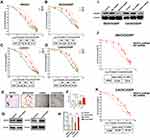
Since the expression of VEGFA correlated with chemotherapy resistance, we further detected its expression in SKOV3, CAOV3, and their DDP resistant cells. Tumor cells were treated with different concentrations of DDP for 24, 48, and 72 hours, and the IC50 of DPP-induced toxicity assessed. The DDP resistant cells were found to have a higher survival percentage than parental tumor cells. The IC50 values of SKOV3 were 27.76, 15.16, and 8.13µM for 24, 48, and 72 hours respectively. SKOV3/DDP had IC50 values of 41.51, 27.19, and 15.22 µM for 24, 48, and 72 hours, respectively. The IC50 values for 24, 48, and 72 hours were 22.49, 20.9, and 8.172 µM, respectively for CAOVE and 33.99, 30.32, and 15.83 µM for CAOV3/DDP (Figure 3A–D). This confirmed that DDP resistant tumor cells were tolerant to DDP killing. Additionally, DDP-resistant cell had more stronger ability to form spheres under DDP treatment (Figure 3E and F). Western blot analysis revealed a high expression of VEGFA in DDP resistant cells than their parental tumor cells (Figure 3G and H). To further investigate the role of VEGFA in chemotherapy resistance, we knocked down VEGFA in SKOV3/DDP and CAOV3/DDP tumor cells. The knockdown of VEGFA significantly reduced the protein level of VEGFA (Figure 3I) and promoted killing sensitivity to chemotherapy drugs. The DPP IC50 values were significantly lower in shVEGFA DDP resistant tumor cells than in control tumor cells following 48 hours of drug treatment (shVEGFA SKOV3/DDP: 12.82 vs. NC: 23.4; shVEGFA CAOV3/DDP: 12.23 vs. NC: 37.01µM, respectively) (Figure 3J and K). These findings suggested that VEGFA promoted chemotherapy resistance of OvCa cells.
Chemotherapy Drugs Induce Autophagy in DDP-Resistant Cells
Several chemotherapeutic drugs have been reported to induce autophagy.21–23 This has been proposed to be a cytoprotective response of cancer cells to cope with the cellular stress imposed by anti-cancer drug treatment. To determine whether autophagy was involved in chemotherapy resistance, the expression of characterized autophagy biomarkers was compared between SKOV3 and SKOV3/DDP as well as CAOV3 and CAOV3/DDP cells. Higher levels of Beclin-1 and LC3-II and lower levels of p62 were observed in SKOV3/DDP and CAOV3/DDP tumor cells than in SKOV3 and CAOV3 cells. This suggested that autophagy was more active in chemotherapy resistant tumor cells (Figure 4A). Next, we examined the expression of these autophagy-related proteins following SKOV3 treatment with DDP (50µM), PTX (40µM), and ADM (20 µM) for 48 h (Figure 4B). The results revealed increased levels of Beclin-1 and LC3-II lipidation, confirming that the chemotherapy drugs could also induce autophagy. To assess whether the expression of VEGFA in OvCa cells induced autophagy, the expression of the autophagy biomarkers in SKOV3/DDP cells was assessed by VEGFA knockdown, autophagy inhibitor 3-MA as a positive control. The knockdown of VEGFA decreased LC3B-II and Beclin1 levels and increased p62 levels (Figure 4C). To further explore the mechanisms that VEGFA mediate DDP-resistance, we detected both pre and cleaved caspase3 expression. We found that knocking down VEGFA increased the expression of cleaved caspase3 in tumor cells. This effect can be reversed by the addition of 3-MA, while, when addition of bafilomycin A1 increased LC3II expression, while, no difference was detected in cleaved-caspase3 expression, indicating that VEGFA may promote the formation of autophagosomes. Next, we added recombinant VEGFA protein in shVEGFA SKOVE/DDP cells, we found that the addition of recombinant VEGFA decreased cleaved-caspase3 expression and increased LC3II expression (Figure 4D). We further examined the effects of VEGFA knockdown and 3MA treatment on SKOV3/DPP cell viability through the assessment of Annexin V and PI staining (Figure 4E). VEGFA knockdown increased the number of early-, late- and total- apoptotic cells (Figure 4F). Moreover, VEGFA knockdown was a more potent inducer of SKV03/DPP cell apoptosis than 3-MA treatment, as evidenced by the higher levels of total apoptosis. This indicated that VEGFA mediated chemotherapy resistance is partly through autophagy. These results confirmed the roles for both VEGFA and autophagy in the survival of SKV03/DPP cells. The VEGFA expression protects tumor cells from chemotherapy killing by stimulating autophagy. Therefore, targeting VEGFA could be a potential therapeutic strategy to enhance chemotherapy treatment in ovarian cancer patients.
Discussion
The overexpression of VEGFA in OvCa has previously been reported and is not limited to gynecological cancers.24–27 However, the mechanisms by which VEGFA promotes tumorigenesis and cancer cell survival remained controversial, most likely due to its tissue-specific roles. Recurrence and drug resistance in OvCa are frequent, but the cellular mechanisms of chemotherapy resistance were poorly characterized.
This study has proved that VEGFA is highly expressed in OvCa tumor tissue and cell lines compared to normal tissue and immortalized ovarian surface epithelium cells at the mRNA and protein levels. Higher levels of VEGFA expression were observed in drug-resistant OvCa cells than in non-drug resistant cells. The chemotherapy drugs, including DDP, PTX, and AMD, elevated the expression of VEGFA. Therefore, VEGFA expression could be an adaptive response employed by OvCa cells to protect themselves from chemotherapy-induced genotoxic stress. Silencing VEGFA increased the potency of DDP to induce OvCa cytotoxicity in SKOV3/DDP and A2780/DDP cells significantly, hence confirming its role in DDP-resistance. The VEGFA mediated drug resistance is in-part, mediated by its ability to regulate autophagy induction. Thus, our findings revealed that inhibition of VEGFA could improve chemotherapy sensitivity in DPP resistant OvCa cells.
The overexpression of VEGFA in OvCa has previously been reported and is not limited to gynecological cancers.24–27 However, the mechanisms by which VEGFA promotes tumorigenesis and cancer cell survival have remained controversial most likely due to its tissue-specific roles. Recurrence and drug resistance in OvCa are common, but the cellular mechanisms of chemotherapy resistance still remain poorly characterized. The VEGFA is upregulated by OvCa cells as a response to the genotypic stress induced by chemotherapeutic agents to promote cell survival. We observed that VEGFA knockdown in chemotherapy-resistant OvCa cells restored the toxicity of chemotherapy compounds. This indicates that sustained VEGFA upregulation is an active survival process, as opposed to a general response to chemotherapy-induced cell shock. A higher expression of VEGFA was observed in malignant tumor cells.28 Yao et al noted that VEGFA mRNA stability is regulated by numerous effectors, including hypoxia-induced growth factors that bind to stabilizing and destabilizing proteins at AU-rich elements in the 3ʹ-UTR of VEGFA. Furthermore, many microRNAs (miRNAs) regulate the expression of VEGFA mRNA in many cancers. For instance, miR-503, miR-497, and miR-185 regulate the expression of VEGFA mRNA in prostate cancer, hepatocellular carcinoma, and clear cell renal cell carcinoma, respectively.9,29,30 Further understanding of VEGFA transcriptional regulation in OvCa is required to reveal the mechanism(s) of enhanced VEGFA in OvCa and its response to chemotherapy drugs. This will allow for the discovery of new strategies to suppress VEGFA expression in OvCa tumors and potentially restoring their sensitivity to chemotherapeutic agents.
The treatment of chemotherapy-resistant-OvCa cells with chemotherapy drugs led to increased autophagy induction. We supposed that the ability of VEGFA to confer protection to OvCa cells was through this pathway since the knockdown of VEGFA inhibited autophagy and led to enhanced OvCa cell apoptosis. Autophagy frequently occurs during tumorigenesis and cancer chemotherapy and is generally believed to confer protection to cancer cells, leading to cancer drug resistance. However, the role of autophagy in cancer development is more complicated. It can be protective for cancer through the removal of damaged organelles and the recycling of degradation products under stress-induced scenarios such as the hypoxic environment of a tumor and drug-induced insult.31 Paradoxically, excessive autophagy can induce cancer cells to undergo autophagic cell death and/or apoptosis.23 Autophagy plays a dual role in the resistance of cancer cells to chemotherapy. It is activated as a protective mechanism to mediate Multi-Drug Resistance (MDR) during drug treatment, and as such, the inhibition of autophagy can re-sensitize resistant cancer cells to chemotherapeutic agents. Secondly, autophagy activation can confer a protective phenotype during drug treatment that prevents programmed cell death.
The role of autophagy in tumor cells is likely to be dependent on both tumor type and stage, and thus must be considered in the context of each cancer.32 We found that in OvCa cells, VEGFA knockdown reduced LC3-II formation and p62 degradation, and thus suppressed autophagy. However, the role of p62 in DDP resistance is controversial. Yu et al reported that SKOV3/DDP cells have much higher p62 levels, and binds to ubiquitinated proteins for transport to autophagic degradation, resulting DDP resistance.33 But, research results are opposite, Zhang et al found that DDP treatment decreased p62 expression.34,35 P62 has a multifunction in regulating survival and death of tumor cells according to the stress of the environment,36 in our study, SKOVE/DDP and CAOV3/DDP cells were maintained with in a low concentration of DDP, this may cause the difference of observation. The DDP-resistance OvCa lines displayed sustained and elevated levels of both VEGFA and autophagy biomarkers, signifying the importance of this pathway in OvCa drug resistance. This study demonstrated the ability of VEGFA to confer OvCa cell protection through sustained autophagy induction. However, the mechanisms by which VEGFA regulates autophagy warrant further investigation.
Inhibiting autophagy in OvCA tumors could represent a viable strategy to alleviate chemotherapy resistance. Numerous studies have demonstrated that autophagy can be inhibited pharmacologically through targeting the early or late stages of the autophagic process.8 Inhibitors that target the early stages include 3-MA, wortmannin, and LY294002, which inhibit the class III PI3Kinase (Vps34).37 These pathways appear to be intact in OvCa tumors because we illustrated the ability of 3-MA to inhibit autophagy and induce apoptosis in SKOV3/DPP cells in this study. Based on targeting of autophagic survival to promote the efficacy of anti-cancer therapy, several clinical trials were conducted in which inhibitors of autophagy were assessed for their ability to benefit renal, prostate, and breast cancer patients.38 The findings of this study suggest that similar trials should be launched in OvCa patients to improve chemotherapy effectiveness and patient prognosis.
Conclusions
In conclusion, our results show that VEGFA expression is associated with chemotherapy resistance and is induced by chemotherapy drugs. Knocking down VEGFA in DDP-resistance OvCa cells enhances drug sensitivity partly by inhibiting autophagy and inducing cleaved caspase3 expression. This suggests that VEGFA may be a novel OvCa therapeutic target. Given the complexity of VEGFA alternative splicing to generate different isoforms with varying bioavailability, future studies should focus on the identification of the specific VEGFA isoform(s) conferring OvCa drug resistance. Emerging Next-Generation-Sequencing (NGS) technologies that can rapidly profile the molecular basis of OvCa tumors make these experiments feasible. Interventions that can inhibit VEGFA activity and impede its ability to maintain OvCa autophagy are needed to restore the effectiveness of chemotherapy treatments during OvCa therapy.
Data Sharing Statement
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
This work was supported by the Basic and Advanced Technology Research Foundation from Science and Technology Department of Henan Province (Grant No.172102310008).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Zeng C, Chen T, Zhang Y, et al. Hedgehog signaling pathway regulates ovarian cancer invasion and migration via adhesion molecule CD24. J Cancer. 2017;8(5):786–792.
2. Balasubramaniam S, Beaver JA, Horton S, et al. FDA approval summary: rucaparib for the treatment of patients with deleterious BRCA mutation-associated advanced ovarian cancer. Clin Cancer Res. 2017;23(23):7165–7170. doi:10.1158/1078-0432.CCR-17-1337
3. Eoh KJ, Yoon JW, Lee JY, et al. A novel algorithm for the treatment strategy for advanced epithelial ovarian cancer: consecutive imaging, frailty assessment, and diagnostic laparoscopy. BMC Cancer. 2017;17(1):481. doi:10.1186/s12885-017-3476-1
4. Fleming ND, Coleman RL, Tung C, et al. Phase II trial of bevacizumab with dose-dense paclitaxel as first-line treatment in patients with advanced ovarian cancer. Gynecol Oncol. 2017;147(1):41–46. doi:10.1016/j.ygyno.2017.07.137
5. Pchejetski D, Alfraidi A, Sacco K, et al. Histone deacetylases as new therapy targets for platinum-resistant epithelial ovarian cancer. J Cancer Res Clin Oncol. 2016;142(8):1659–1671. doi:10.1007/s00432-015-2064-5
6. Bookman MA. First-line chemotherapy in epithelial ovarian cancer. Clin Obstet Gynecol. 2012;55(1):96–113. doi:10.1097/GRF.0b013e31824b45da
7. Jang K, Kim M, Gilbert CA, et al. VEGFA activates an epigenetic pathway upregulating ovarian cancer-initiating cells. EMBO Mol Med. 2017;9(3):304–318. doi:10.15252/emmm.201606840
8. Wang W, Ren F, Wu Q, et al. MicroRNA-497 suppresses angiogenesis by targeting vascular endothelial growth factor A through the PI3K/AKT and MAPK/ERK pathways in ovarian cancer. Oncol Rep. 2014;32(5):2127–2133. doi:10.3892/or.2014.3439
9. Yan JJ, Zhang YN, Liao JZ, et al. MiR-497 suppresses angiogenesis and metastasis of hepatocellular carcinoma by inhibiting VEGFA and AEG-1. Oncotarget. 2015;6(30):29527–29542. doi:10.18632/oncotarget.5012
10. Zeng FC, Zeng MQ, Huang L, et al. Downregulation of VEGFA inhibits proliferation, promotes apoptosis, and suppresses migration and invasion of renal clear cell carcinoma. Onco Targets Ther. 2016;9:2131–2141. doi:10.2147/OTT.S98002
11. Zhang W, Zou C, Pan L, et al. MicroRNA-140-5p inhibits the progression of colorectal cancer by targeting VEGFA. Cell Physiol Biochem. 2015;37(3):1123–1133. doi:10.1159/000430237
12. Lawicki S, Bedkowska GE, Gacuta-Szumarska E, et al. The plasma concentration of VEGF, HE4 and CA125 as a new biomarkers panel in different stages and sub-types of epithelial ovarian tumors. J Ovarian Res. 2013;6(1):45. doi:10.1186/1757-2215-6-45
13. Fahmy K, Gonzalez A, Arafa M, et al. Myoferlin plays a key role in VEGFA secretion and impacts tumor-associated angiogenesis in human pancreas cancer. Int J Cancer. 2016;138(3):652–663. doi:10.1002/ijc.29820
14. Borst P, Rottenberg S, Jonkers J. How do real tumors become resistant to cisplatin? Cell Cycle. 2008;7(10):1353–1359. doi:10.4161/cc.7.10.5930
15. Weckman A, Rotondo F, Di Ieva A, et al. Autophagy in endocrine tumors. Endocr Relat Cancer. 2015;22(4):R205–R218. doi:10.1530/ERC-15-0042
16. Takahashi N, Iwasa S, Taniguchi H, et al. Prognostic role of ERBB2, MET and VEGFA expression in metastatic colorectal cancer patients treated with anti-EGFR antibodies. Br J Cancer. 2016;114(9):1003–1011. doi:10.1038/bjc.2016.74
17. Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
18. Barretina J, Caponigro G, Stransky N, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi:10.1038/nature11003
19. Ren XL, He GY, Li XM, et al. MicroRNA-206 functions as a tumor suppressor in colorectal cancer by targeting FMNL2. J Cancer Res Clin Oncol. 2016;142(3):581–592. doi:10.1007/s00432-015-2053-8
20. Lu W, Wang X, Liu J, et al. Downregulation of ARHGDIA contributes to human glioma progression through activation of Rho GTPase signaling pathway. Tumour Biol. 2016;37(12):15783–15793. doi:10.1007/s13277-016-5374-6
21. Guo X-L, Li D, Hu F, et al. Targeting autophagy potentiates chemotherapy-induced apoptosis and proliferation inhibition in hepatocarcinoma cells. Cancer Lett. 2012;320(2):171–179. doi:10.1016/j.canlet.2012.03.002
22. Ramakrishnan R, Huang C, Cho H-I, et al. Autophagy induced by conventional chemotherapy mediates tumor cell sensitivity to immunotherapy. Cancer Res. 2012;72(21):5483–5493. doi:10.1158/0008-5472.CAN-12-2236
23. Ropolo A, Bagnes CI, Molejon MI, et al. Chemotherapy and autophagy-mediated cell death in pancreatic cancer cells. Pancreatology. 2012;12(1):1–7. doi:10.1016/j.pan.2011.11.003
24. Aktas SH, Akbulut H, Yazici O, et al. A new angiogenesis prognostic index with VEGFA, PlGF, and angiopoietin1 predicts survival in patients with advanced gastric cancer. Turk J Med Sci. 2017;47(2):399–406. doi:10.3906/sag-1509-80
25. Baba H, Baba Y, Uemoto S, et al. Changes in expression levels of ERCC1, DPYD, and VEGFA mRNA after first-line chemotherapy of metastatic colorectal cancer: results of a multicenter study. Oncotarget. 2015;6(32):34004–34013. doi:10.18632/oncotarget.5227
26. Braicu EI, Gasimli K, Richter R, et al. Role of serum VEGFA, TIMP2, MMP2 and MMP9 in monitoring response to adjuvant radiochemotherapy in patients with primary cervical cancer–results of a companion protocol of the randomized NOGGO-AGO Phase III clinical trial. Anticancer Res. 2014;34(1):385–391.
27. Chen B, Zhang C, Dong P, et al. Molecular regulation of cervical cancer growth and invasion by VEGFa. Tumour Biol. 2014;35(11):11587–11593. doi:10.1007/s13277-014-2463-2
28. Yao P, Potdar AA, Ray PS, et al. The HILDA complex coordinates a conditional switch in the 3ʹ-untranslated region of the VEGFA mRNA. PLoS Biol. 2013;11(8):e1001635. doi:10.1371/journal.pbio.1001635
29. Ma X, Shen D, Li H, et al. MicroRNA-185 inhibits cell proliferation and induces cell apoptosis by targeting VEGFA directly in von Hippel-Lindau-inactivated clear cell renal cell carcinoma. Urol Oncol. 2015;33(4):169 e161–111. doi:10.1016/j.urolonc.2015.01.003
30. Zhu S, Cao L, Yu Y, et al. Inhibiting autophagy potentiates the anticancer activity of IFN1@/IFNalpha in chronic myeloid leukemia cells. Autophagy. 2013;9(3):317–327. doi:10.4161/auto.22923
31. Gong C, Song E, Codogno P, et al. The roles of BECN1 and autophagy in cancer are context dependent. Autophagy. 2012;8(12):1853–1855. doi:10.4161/auto.21996
32. Dikic I, Johansen T, Kirkin V. Selective autophagy in cancer development and therapy. Cancer Res. 2010;70(9):3431–3434. doi:10.1158/0008-5472.CAN-09-4027
33. Yu H, Su J, Xu Y, et al. p62/SQSTM1 involved in cisplatin resistance in human ovarian cancer cells by clearing ubiquitinated proteins. Eur J Cancer. 2011;47(10):1585–1594. doi:10.1016/j.ejca.2011.01.019
34. Liu B, Qi X, Zhang X, et al. Med19 is involved in chemoresistance by mediating autophagy through HMGB1 in breast cancer. J Cell Biochem. 2019;120(1):507–518. doi:10.1002/jcb.27406
35. Zhang X, Wang LL, Wang B, et al. Effect of siRNA-induced Atg7 gene silencing on the sensitivity of ovarian cancer SKOV3 cells to cisplatin. Am J Transl Res. 2020;12(5):2052–2061.
36. Yan XY, Qu XZ, Xu L, et al. Insight into the role of p62 in the cisplatin resistant mechanisms of ovarian cancer. Cancer Cell Int. 2020;20:128. doi:10.1186/s12935-020-01196-w
37. Liang B, Kong D, Liu Y, et al. Autophagy inhibition plays the synergetic killing roles with radiation in the multi-drug resistant SKVCR ovarian cancer cells. Radiat Oncol. 2012;7:213. doi:10.1186/1748-717X-7-213
38. Yang ZJ, Chee CE, Huang S, et al. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther. 2011;10(9):1533–1541. doi:10.1158/1535-7163.MCT-11-0047
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.