Back to Journals » OncoTargets and Therapy » Volume 13
Inhibition of cyclinB1 Suppressed the Proliferation, Invasion, and Epithelial Mesenchymal Transition of Hepatocellular Carcinoma Cells and Enhanced the Sensitivity to TRAIL-Induced Apoptosis
Authors Lv S, Ning H, Li Y, Wang J, Jia Q, Wen H
Received 29 July 2019
Accepted for publication 15 November 2019
Published 5 February 2020 Volume 2020:13 Pages 1119—1128
DOI https://doi.org/10.2147/OTT.S225202
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Nicola Silvestris
Shuai Lv, Hanbing Ning, Yingxia Li, Jingyun Wang, Qiaoyu Jia, Hongtao Wen
Department of Gastroenterology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450018, Henan Province, People’s Republic of China
Correspondence: Shuai Lv; Hongtao Wen
Department of Gastroenterology, The First Affiliated Hospital of Zhengzhou University, No. 1 East Jianshe Road, Zhengzhou 450018, Henan Province, People’s Republic of China
Tel +86-371-6627-1112
Email [email protected]; [email protected]
Background: CyclinB1 is highly expressed in various tumor tissues and plays an important role in tumor progression. However, its role in hepatocellular carcinoma (HCC) remains unclear. Therefore, the aim of this study was to explore the role of cyclinB1 in the development and progression of HCC.
Methods: The expression of cyclinB1 was analyzed using the Gene Expression Profiling Interactive Analysis (GEPIA) database, and detected in HCC tissues and HCC cell lines through quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and Western blotting. CyclinB1-short hairpin RNA (Sh-cyclinB1) was transfected into HCC cells to knockdown cyclinB1, and the effect of cyclinB1 knockdown on HCC was examined via the MTT assay, colony formation assay, wound healing assay, scratch assay, cell cycle analysis in vitro, and xenograft model in nude mice. In addition, the role of cyclinB1 on tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis was measured using flow cytometry and Western blotting.
Results: The GEPIA database analysis showed that cyclinB1 was highly expressed in HCC tissues. The results of qRT-PCR and Western blotting proved that the expression of cyclinB1 was significantly increased in HCC tissues and cell lines. The data of the MTT assay, colony formation assay, and cell cycle analysis indicated that cyclinB1 knockdown inhibited the proliferation of HCC cells. In addition, cell migration, invasion, and epithelial mesenchymal transition were also impaired by cyclinB1 knockdown. Furthermore, the xenograft model in nude mice demonstrated that inhibition of cyclinB1 suppressed tumor growth and metastasis in vivo. Finally, the results of flow cytometry and Western blotting indicated that inhibition of cyclinB1 enhanced the sensitivity of HCC cells to TRAIL-induced apoptosis.
Conclusion: Overall, these data provide reasonable evidence that cyclinB1 may serve as a proto-oncogene during the progression of HCC.
Keywords: cyclinB1, proliferation, hepatocellular carcinoma, TRAIL
Introduction
Primary liver cancer is one of the main causes of cancer-related death globally, and remains one of the most refractory types of cancer.1 Hepatocellular carcinoma (HCC) is the most common type of liver cancer, accounting for approximately 90% of primary liver cancer cases.2 HCC accounts for approximately 4–5% of newly diagnosed cancers worldwide, and approximately 560,000 individuals are diagnosed with HCC annually.3 In China, the risk of HCC is particularly high, accounting for approximately 55% of cases worldwide.4 In spite of the advances in diagnosis and treatment (e.g., surgical excision, transplantation, chemotherapy, and drug therapy), the survival rate remains unfavorable and the 5-year survival rate has not significantly improved.5 In this regard, uncovering novel biomarkers for early diagnosis and new targets for molecular therapy is imperative to improve the survival rate of patients with HCC.
CyclinB1 and activation of the cyclin-dependent kinase 1/cyclinB1 complex have been shown to regulate the cell cycle transition from G2 to the M phase.6,7 Increased expression of cyclinB1 accelerates mitosis and may lead to excessive proliferation of cells. Studies have shown that cyclinB1 is highly expressed in various tumor tissues, and plays an important role in tumor growth. A multivariate analysis demonstrated that overexpression of cyclinB1 may be related to the malignant development of laryngeal squamous cell carcinoma.8 A study demonstrated that tumor tissues expressing high levels of cyclinB1 were resistant to radiotherapy, and the expression level of cyclinB1 is likely to be an important indicator of the risk of recurrence and metastasis in patients with head and neck squamous cell carcinoma after radiotherapy.9 Overexpression of cyclinB1 was also observed in oral squamous cell carcinoma, and was involved in early carcinogenesis, cell differentiation, and tumor proliferation.10 However, the expression and specific function of cyclinB1 in HCC remains unclear.
Therefore, we explored the specific role of cyclinB1 in the pathogenesis of HCC. We examined the expression level of cyclinB1 in HCC through quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and immunohistochemistry staining of HCC specimens. In addition, short hairpin RNA-mediated (shRNA-mediated) gene knockdown was applied to explore the specific biological functions of cyclinB1 in HCC cell lines. The role of cyclinB1 on tumor necrosis factor-related apoptosis-inducing ligand-induced (TRAIL-induced) apoptosis was also investigated.
Materials and Methods
Tissue Sample Collection
HCC tissues and adjacent normal tissues were obtained from 28 patients who underwent tumor resection in the First Affiliated Hospital of Zhengzhou University (Zhengzhou, China) from May 2016 to November 2018. The use of tumor samples in this study was approved by the ethics committee of the First Affiliated Hospital of Zhengzhou University. All patients provided written informed consent.
Cell Lines and cell Culture
The HCC cell lines HepG2, BEL-7402, Huh-7, Hep3B, SMCC7721, and MHCC97H, as well as the normal human liver cell line LO-2 were used in this study. BEL-7402, Huh-7, and Hep3B cell were obtained from the Library of Typical Culture of Chinese Academy of Sciences (Shanghai, China). MHCC97H and SMCC7721 cells were purchased from the Liver Cancer Institute of Fudan University (Shanghai, China). HepG2 and LO-2 cells and purchased from the American Type Culture Collection (Manassas, VA, USA). All cells were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen, Waltham, MA, USA) with 10% fetal bovine serum (Invitrogen, USA) and 1% penicillin/streptomycin. The cells were cultured in an incubator at 37°C in a humidification atmosphere containing 5% CO2.
Knockdown of cyclinB1
Specific shRNA sequence targeting cyclinB1 messenger RNA (mRNA) sequence or a control shRNA was cloned into the pLKO.1-puro vector (Sigma) to stably knock down the expression of cyclinB1 in HCC cells. Subsequently, Lipofectamine 2000 reagent (Invitrogen) was used to transfect the vector into HCC cells according to the instructions provided by the manufacturer.
RNA Extraction and Real-Time PCR
Total RNA from tissue samples and cells was isolated using the Trizol reagent (Invitrogen) following the protocol provided by the manufacturer. The PrimeScriptTM RT regent Kit with gDNA Eraser (Takara, Tokyo, Japan) was employed for the removal of genomic DNA and qRT-PCR. Denaturation, followed by 40 cycles of PCR reactions and a dissociation stage were performed on a CFX Connect™ Real-Time PCR Detection System (Takara). The relative expression level of cyclinB1 was calculated using the 2−ΔΔCt method and normalized to that of β-actin. The experiment was performed thrice and each sample was assayed in triplicate.
Western Blotting
Cells were lysed with radioimmunoprecipitation assay buffer containing complete protease inhibitor (Sigma). The Bradford assay was used for quantification. An equal amount of protein was used in each group for subsequent analysis. Western blotting of proteins was performed according to standard experimental procedures. The following antibodies were used in the Western blotting analysis: rabbit anti-cyclinB1 antibody (1:5000, ab32053; Abcam), mouse anti-vimentin antibody (1:500, ab8978; Abcam), rabbit anti-N-Cadherin antibody (1:1000, ab18203; Abcam), mouse anti-E-Cadherin antibody (1:50, ab1416; Abcam), anti-cleaved caspase-3 (1 µg/mL, ab2302; Abcam), and anti-cleaved caspase-9 (1 µg/mL, ab2324; Abcam).
MTT Assay
Cell viability after cyclinB1 knockdown was measured through the MTT assay. Following transfection of Huh-7 and HepG2 cells with sh-cyclinB1 or negative control-shRNA (sh-NC) for 72 h, cells were planted in a 96-well plate at a density of 5000 cells per well. At the indicated timepoint, MTT (20 μL, 5 mg/mL; Sigma−Aldrich) solution was added and the cells were incubated for 4 h. After centrifugation, the supernatant was discarded and 200 μL dimethyl sulfoxide (DMSO) was added. The absorbance was measured at 570 nm using a microplate reader. The experiment was performed thrice and each sample was assayed in five replicates.
Colony Formation Assay
Cells at the logarithmic growth stage were digested with 0.25% trypsin inoculated into a dish containing 10 mL culture medium preheated to 37°C and gently rotated to disperse the cells evenly. The cells were incubated at 37°C with 5% CO2 and saturated humidity for 2 weeks. The culture was terminated following the appearance of visible clones. The supernatant was discarded and the cells were gently washed twice with phosphate-buffered saline (PBS). The cells were fixed with 4% paraformaldehyde for 15 min and stained with Giemsa stain (Solarbio, Beijing, China) for 30 min. Subsequently, the rate of clone formation was calculated.
Apoptosis Assay
The apoptosis ratio was analyzed using the Annexin V-fluorescein isothiocyanate Apoptosis Detection Kit (Beyotime, China). At 48 h after plasmid transfection, HCC cells were harvested, washed with cold PBS, resuspended in working solution containing Annexin binding buffer and propidium iodide stock solution according to the manufacturer’s instructions. After incubation for 15 min at room temperature in the dark, samples were analyzed using the Muse Cell Analyzer (Millipore, Hayward, CA, USA) and MuseSoft 1.4.0.0 (Millipore).
Transwell Invasion Assay
The invasive ability of HCC cells transfected with sh-cyclinB1 or sh-NC was evaluated in a 24‑well Matrigel Invasion Chamber (BD Biosciences, San José, CA, USA). A total of 1×105 cells per mL were suspended in serum-free medium containing bovine serum albumin, cell suspension (200 μL) was added to the upper chamber, and medium containing 5% fetal bovine serum (500 μL) was added to the lower chamber. After incubation for 24 h, the noninvasive cells which remained in the upper chamber were removed using a clean cotton swab, whereas the invasive cells in the lower chamber were fixed with 3.7% paraformaldehyde. The cells were washed with PBS, stained with hematoxylin at room temperature for 1 h, and visualized with a microscope.
Wound Healing Assay
The cells were inoculated into six-well plates at a density of 90% and incubated overnight in an incubator. A scratch perpendicular to the cell layer was performed using a 200-μL pipette. Subsequently, cells were rinsed twice with PBS to remove loose cells, and fresh medium was added to continue the culture. Photographs were captured at 0 h and 24 h to assess cell migration to the wound.
Xenograft Model in Nude Mice
Six-week-old BALB/c nude mice were purchased from Shanghai Super-B&K Laboratory Animal Corp., Ltd. (Shanghai, China). HCC cells transfected with sh-cyclinB1 or sh-NC were centrifuged and suspended in DMEM medium. Subsequently, 1×107 cells suspended in 200 μL culture medium were subcutaneously injected into the outer hips of each mouse. The mice were cultured in a pathogen-free environment with adequate water and food for 28 days. Tumor growth was monitored every 7 days. The mice were finally sacrificed, and the tumor volumes were calculated according to the following formula: V = a×b×(a+b)/2. The animal care and experimental protocols were approved by the Animal Care Committee of the First Affiliated Hospital of Zhengzhou University, and were in accordance with Henan Province Laboratory Animal Care Guidelines for the use of animals.
Statistical Analysis
Data were represented as mean ± standard deviation of three independent experiments, and the SPSS 17.0 statistical software (SPSS, Chicago, IL, USA) was used for processing. A Student paired t-test was used to compare the differences between the two groups. A p<0.05 denoted statistical significance.
Results
CyclinB1 Was Upregulated in HCC Tissues and Cell Lines
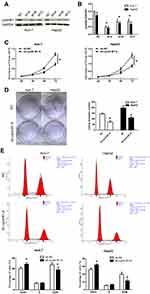
Firstly, the expression of cyclinB1 in HCC was explored using the Gene Expression Profiling Interactive Analysis database (http://gepia.cancer-pku.cn/index.html) to identify the role of cyclinB1 in HCC. As shown in Figure 1A, the expression of cyclinB1 was significantly higher in liver HCC tissues (n = 369) than normal liver tissues (n = 160).
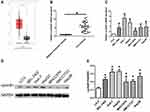 |
Figure 1 CyclinB1 was upregulated in HCC tissues and cell lines. (A) The expression of cyclinB1 in liver hepatocellular carcinoma (LIHC) was explored using the GEPIA database (http://gepia.cancer-pku.cn/index.html). (B) HCC tissues and the paired adjacent normal tissues were collected from patients with HCC. qRT-PCR was performed to quantify the mRNA expression of cyclinB1. *p<0.05 vs. adjacent normal tissues. (C) qRT-PCR was performed to quantify the mRNA expression of cyclinB1 in HCC cell lines BEL-7402, Huh-7, HepG2, MHCC97H, SMCC7721, and Hep3B, as well as the normal liver cell line LO-2. *p<0.05 vs. LO-2. (D, E) Western blotting was performed to detect the protein expression of cyclinB1 in HCC cell lines BEL-7402, Huh-7, HepG2, MHCC97H, SMCC7721, and Hep3B, as well as the normal liver cell line LO-2. |
Secondly, the expression of cyclinB1 in HCC tissues and their paired adjacent nontumor tissues obtained from 28 patients was examined through qRT-PCR. The results showed that the expression level of cyclinB1 was notably increased in HCC tissues compared with adjacent nontumor tissues (Figure 1B). Simultaneously, the expression of cyclinB1 in the HCC cell lines BEL-7402, Huh-7, HepG2, MHCC97H, SMCC7721, Hep3B, and the normal liver cell line LO-2 were detected using qRT-PCR and Western blotting. We observed that both the mRNA and protein expression levels of cyclinB1 were higher in HCC cell lines than those measured in normal liver cell lines (Figure 1C–E).
Knockdown of cyclinB1 Restrained Proliferation of HCC Cells
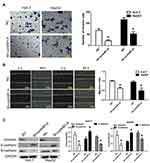
Huh-7 and HepG2 cells showed the highest expression of cyclinB1; thus, they were employed for the following experiments. These cells were transfected with sh-cyclinB1-A, sh-cyclinB1-B, and sh-cyclinB1-C to knock down cyclinB1, and scramble shRNA was used as the NC. As indicated by Figure 2A and B, sh-cyclinB1-A resulted in the lowest expression of cyclinB1. Hence, sh-cyclinB1-A was selected and transfected into Huh-7 and HepG2 cells to stably silence cyclinB1. MTT assay and colony formation assay were conducted to explore the potential role of cyclinB1 on the proliferation of HCC cells. The results of the MTT assay showed that inhibition of cyclinB1 in Huh-7 and HepG2 cells restrained cell viability compared with the NC group (Figure 2C). Similarly, the results of the colony formation assay demonstrated that knockdown of cyclinB1 reduced the number of formed clones (Figure 2D). In addition, cell cycle arrest in Huh-7 and HepG2 cells was detected through flow cytometry. The data demonstrated that knockdown of cyclinB1 in Huh-7 and HepG2 cells notably increased the percentage of cells in the G0/G1 phase, whereas it decreased that of cells in the G2/M phase (Figure 2E).
Knockdown of cyclinB1 Inhibited the Migration and Invasion of HCC Cells and Suppressed Epithelial Mesenchymal Transition (EMT)
A scratch assay and transwell invasion assay were performed to clarify the effect of cyclinB1 on the migration and invasion of HCC cells. As shown by the transwell invasion assay, the invasive ability of Huh-7 and HepG2 cells significantly declined upon knockdown of cyclinB1 (Figure 3A). Consistently, the migratory ability of Huh-7 and HepG2 cells was significantly decreased in the sh-cyclinB1-A group compared with that recorded in the NC group (Figure 3B). Next, we investigated whether cyclinB1 exerts a regulatory effect on EMT. The expression of EMT-associated proteins vimentin, N-cadherin, and E-cadherin was investigated via Western blotting. The results showed that knockdown of cyclinB1 in Huh-7 and HepG2 cells restrained the expression of vimentin and N-cadherin, while that of E-cadherin was enhanced compared with the NC group (Figure 3C).
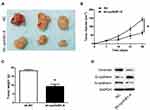
Knockdown of cyclinB1 Inhibited Tumor Growth in the Xenograft Model of Nude Mice
We established a xenograft tumor model in nude mice to investigate whether knockdown of cyclinB1 can inhibit the development and progression of tumors in vivo. Subcutaneous injection of Huh-7 cells transfected with sh-cyclinB1-A- or NC was performed on the lateral gluteal surface of nude mice. As expected, we observed that knockdown of cyclinB1 significantly suppressed tumor formation compared with the NC group (Figure 4A). Of note, the tumor volume and weight were both markedly reduced in the cyclinB1 knockdown group after 4 weeks (Figure 4B and C). Furthermore, knockdown of cyclinB1 restrained the expression of vimentin and N-cadherin, whereas that of E-cadherin was enhanced compared with the NC group (Figure 4D).
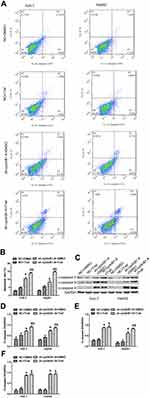
Knockdown of cyclinB1 Sensitized HCC Cells to TRAIL-Induced Apoptosis
We further explored whether knockdown of cyclinB1 has an effect on TRAIL-induced apoptosis. Huh-7 and HepG2 cells transfected with sh-cyclinB1-A or NC were treated with TRAIL or DMSO, and a flow cytometric analysis was performed to detect apoptosis. The results showed that knockdown of cyclinB1 promoted the apoptosis of Huh-7 and HepG2 cells compared with that observed in the NC groups (Figure 5A and B). Moreover, incubation with TRAIL further increased the apoptosis of Huh-7 and HepG2 cells compared with that noted in the cyclinB1 knockdown single treatment group (Figure 5A and B). We monitored the protein expression of cleaved-caspase 3, cleaved-caspase 8, and cleaved-caspase 9 through Western blotting to further confirm the promoting effect of cyclinB1 knockdown on TRAIL-induced apoptosis. As expected, the expression of cleaved-caspase 3, cleaved-caspase 8, and cleaved-caspase 9 was increased after knockdown of cyclinB1 in both the sh-cyclinB1-A+DMSO versus NC+DMSO groups and the sh-cyclinB1-A+TRAIL versus NC+TRAIL groups (Figure 5C–F).
Discussion
Numerous studies have shown that high expression of cyclinB1 in various tumors is related to tumor grade and prognosis of patients. A meta-analysis revealed that overexpression of cyclinB1 was associated with poor survival in patients with most types of solid tumors, suggesting that the expression of cyclinB1 is an important parameter for prognosis in this setting.11 It was found that the expression of cyclinB1 was higher in malignant specimens than in benign cases, suggesting that cyclinB1 can be used as an effective marker for the differentiation of benign and malignant thyroid tumors.12 Patients with high expression of cyclinB1 showed worse overall survival versus that of patients with low expression, and the results of the multivariate Cox regression analysis indicated that cyclinB1 was an independent prognostic factor in pancreatic cancer.13 In this study, through qRT‑PCR and immunohistochemical analyses, we observed that cyclinB1 was highly expressed in HCC tissues than in adjacent nontumor tissues. Moreover, using qRT-PCR and Western blotting, we observed that the expression of cyclinB1 in HCC cell lines was significantly higher than that noted in normal liver cells, indicating that cyclinB1 may be a potential target for clinical diagnosis, as well as an oncogenic factor of HCC.
Studies have illustrated that a specific reduction of the expression of cyclinB1 can inhibit the proliferation of tumor cells. Pan et al certified that downregulation of cyclinB1/cyclin-dependent kinase 1 mediated by inhibition of eukaryotic translation initiation factor 3 subunit D restrained tumorigenesis in renal cell carcinoma by inducing cell cycle arrest at the G2/M phase.14 Similarly, Zhang et al verified that treatment with nagilactone E downregulated the expression of cyclinB1, induced cell cycle arrest at the G2 phase, and suppressed proliferation in non-small cell lung cancer cells.15 Kedinger et al demonstrated that silencing of cyclinB1 through the use of small interfering RNAs (siRNAs) blocked the cell cycle and suppressed the proliferation of melanoma cells. Furthermore, siRNAs against cyclinB1 significantly inhibited the growth and lung metastases of subcutaneous tumors.16 In this study, we found that knockdown of cyclinB1 suppressed cell proliferation, migration, and invasion, reduced the number of cells in the G2/M phase in vitro, and restrained tumor growth in vivo. These results indicated that cyclinB1 may be a new target for the treatment of HCC. Some studies have shown that silencing of cyclinB1 is usually accompanied by inhibition of EMT.17–19 In the present study, we investigated the EMT-related markers; the results showed that knockdown of cyclinB1 reversed the progression of EMT and may affect the metastasis of HCC.
Cell cycle-arrested tumor cells are more sensitive to TRAIL-induced apoptosis.20 A variety of chemotherapeutic agents can block the cell cycle of tumor cells and enhance their sensitivity to TRAIL-induced apoptosis.21–23 Since knockdown of cyclinB1 induced cell cycle arrest, we sought to determine whether it would contribute to TRAIL-induced apoptosis in HCC. Our study certified that knockdown of cyclinB1 sensitized HCC cells to TRAIL-induced apoptosis.
In conclusion, we observed that cyclinB1 was highly expressed in HCC tissues and cell lines. Knockdown of cyclinB1 impaired the proliferation, migration, and invasion of HCC cells, arrested their cell cycle in the G0/G1 phase, and reversed the EMT processes. In addition, knockdown of cyclinB1 sensitized HCC cells to TRAIL-induced apoptosis. Collectively, our data provide convincing evidence for the potential usefulness of cyclinB1 as a drug target in the treatment of HCC.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Yang JD, Roberts LR. Hepatocellular carcinoma: a global view. Nat Rev Gastroenterol Hepatol. 2010;7(8):448–458. doi:10.1038/nrgastro.2010.100
2. Tunissiolli NM, Castanhole-Nunes MMU, Biselli-Chicote PM, et al. Hepatocellular carcinoma: a comprehensive review of biomarkers, clinical aspects, and therapy. Asian Pac J Cancer Prev. 2017;18(4):863–872. doi:10.22034/APJCP.2017.18.4.863
3. Nordenstedt H, White DL, El-Serag HB. The changing pattern of epidemiology in hepatocellular carcinoma. Dig Liver Dis. 2010;42(Suppl 3):S206–S214. doi:10.1016/S1590-8658(10)60507-5
4. Gao J, Xie L, Yang WS, et al. Risk factors of hepatocellular carcinoma–current status and perspectives. Asian Pac J Cancer Prev. 2012;13(3):743–752. doi:10.7314/APJCP.2012.13.3.743
5. Kee KM, Wang JH, Wang CC, Cheng YF, Lu SN. Hepatocellular carcinoma associated with extra-hepatic primary malignancy: its secular change, clinical manifestations and survival. Sci Rep. 2016;6:30156. doi:10.1038/srep30156
6. Gavet O, Pines J. Progressive activation of cyclinB1-Cdk1 coordinates entry to mitosis. Dev Cell. 2010;18(4):533–543. doi:10.1016/j.devcel.2010.02.013
7. Tang L, Zhang Y, Pan H, et al. Involvement of cyclin B1 in progesterone-mediated cell growth inhibition, G2/M cell cycle arrest, and apoptosis in human endometrial cell. Reprod Biol Endocrinol. 2009;7:144. doi:10.1186/1477-7827-7-144
8. Dong Y, Sui L, Watanabe Y, Sugimoto K, Tokuda M. Clinical relevance of cyclin B1 overexpression in laryngeal squamous cell carcinoma. Cancer Lett. 2002;177(1):13–19. doi:10.1016/S0304-3835(01)00770-4
9. Hassan KA, Ang KK, El-Naggar AK, et al. Cyclin B1 overexpression and resistance to radiotherapy in head and neck squamous cell carcinoma. Cancer Res. 2002;62(22):6414–6417.
10. Patil GB, Hallikeri KS, Balappanavar AY, Hongal SG, Sanjaya PR, Sagari SG. Cyclin B1 overexpression in conventional oral squamous cell carcinoma and verrucous carcinoma- A correlation with clinicopathological features. Med Oral Patol Oral Cir Bucal. 2013;18(4):e585–e590. doi:10.4317/medoral.18220
11. Ye C, Wang J, Wu P, Li X, Chai Y. Prognostic role of cyclin B1 in solid tumors: a meta-analysis. Oncotarget. 2017;8(2):2224–2232. doi:10.18632/oncotarget.13653
12. Nar A, Ozen O, Tutuncu NB, Demirhan B. Cyclin A and cyclin B1 overexpression in differentiated thyroid carcinoma. Med Oncol. 2012;29(1):294–300. doi:10.1007/s12032-010-9800-0
13. Zhou L, Li J, Zhao YP, et al. The prognostic value of cyclin B1 in pancreatic cance. Med Oncol. 2014;31(9):107. doi:10.1007/s12032-014-0107-4
14. Pan XW, Chen L, Hong Y, et al. EIF3D silencing suppresses renal cell carcinoma tumorigenesis via inducing G2/M arrest through downregulation of cyclin B1/CDK1 signaling. Int J Oncol. 2016;48(6):2580–2590. doi:10.3892/ijo.2016.3459
15. Zhang LL, Feng ZL, Su MX, et al. Downregulation of cyclin B1 mediates nagilactone E-induced G2 phase cell cycle arrest in non-small cell lung cancer cells. Eur J Pharmacol. 2018;830:17–25. doi:10.1016/j.ejphar.2018.04.020
16. Kedinger V, Meulle A, Zounib O, et al. Sticky siRNAs targeting survivin and cyclin B1 exert an antitumoral effect on melanoma subcutaneous xenografts and lung metastases. BMC Cancer. 2013;13:338. doi:10.1186/1471-2407-13-338
17. Pan Y, Wang B, Yang X, et al. CUL4A facilitates hepatocarcinogenesis by promoting cell cycle progression and epithelial-mesenchymal transition. Sci Rep. 2015;5:17006. doi:10.1038/srep17006
18. Li JP, Yang YX, Liu QL, et al. The pan-inhibitor of Aurora kinases danusertib induces apoptosis and autophagy and suppresses epithelial-to-mesenchymal transition in human breast cancer cells. Drug Des Devel Ther. 2015;9:1027–1062. doi:10.2147/DDDT.S74412
19. Wang S, Li P, SM L, Ling ZQ. Chemoprevention of low-molecular-weight citrus pectin (LCP) in gastrointestinal cancer cells. Int J Biol Sci. 2016;12(6):746–756. doi:10.7150/ijbs.13988
20. Ehrhardt H, Wachter F, Grunert M, Jeremias I. Cell cycle-arrested tumor cells exhibit increased sensitivity towards TRAIL-induced apoptosis. Cell Death Dis. 2013;4:e661. doi:10.1038/cddis.2013.179
21. Dai H, Jiang Y, Luo Y, Bie P, Chen Z. Triptolide enhances TRAIL sensitivity of pancreatic cancer cells by activating autophagy via downregulation of PUM1. Phytomedicine. 2019;62:152953. doi:10.1016/j.phymed.2019.152953
22. Kim JL, Park SH, Jeong S, et al. Sea cucumber (Stichopus japonicas) F2 enhanced TRAIL-induced apoptosis via XIAP ubiquitination and ER stress in colorectal cancer cells. Nutrients. 2019;11(5):1061. doi:10.3390/nu11051061
23. Trivedi R, Maurya R, Mishra DP. Medicarpin, a legume phytoalexin sensitizes myeloid leukemia cells to TRAIL-induced apoptosis through the induction of DR5 and activation of the ROS-JNK-CHOP pathway. Cell Death Dis. 2014;5:e1465. doi:10.1038/cddis.2014.429
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.