Back to Journals » OncoTargets and Therapy » Volume 14
Inhibition of circRNA circVPS33B Reduces Warburg Effect and Tumor Growth Through Regulating the miR-873-5p/HNRNPK Axis in Infiltrative Gastric Cancer
Authors Lu Y, Cheng J, Cai W, Zhuo H, Wu G, Cai J
Received 17 November 2020
Accepted for publication 2 April 2021
Published 12 May 2021 Volume 2021:14 Pages 3095—3108
DOI https://doi.org/10.2147/OTT.S292575
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Arseniy Yuzhalin
Yizhuo Lu,1 Jia Cheng,2 Wangyu Cai,2 Huiqin Zhuo,2 Guoyang Wu,1 Jianchun Cai2
1Department of General Surgery, Zhongshan Hospital Xiamen University, Institute of Gastrointestinal Oncology, School of Medicine, Xiamen University, Xiamen Municipal Key Laboratory of Gastrointestinal Oncology, Xiamen, Fujian, 361004, People’s Republic of China; 2Department of Gastrointestinal Surgery, Zhongshan Hospital Xiamen University, Institute of Gastrointestinal Oncology, School of Medicine, Xiamen University, Xiamen Municipal Key Laboratory of Gastrointestinal Oncology, Xiamen, Fujian, 361004, People’s Republic of China
Correspondence: Jianchun Cai
Department of Gastrointestinal Surgery, Zhongshan Hospital Xiamen University, Institute of Gastrointestinal Oncology, School of Medicine, Xiamen University, Xiamen Municipal Key Laboratory of Gastrointestinal Oncology, Room 203, No. 146, Hubin South Road, Siming District, Xiamen, Fujian Province, 361004, People’s Republic of China
Tel +86 592-2993191
Email [email protected]
Background: Circular RNA VPS33B (circVPS33B) has been revealed to be upregulated in gastric cancer (GC) tissues. However, the role of circVPS33B in infiltrative GC is indistinct.
Methods: Expression of circVPS33B was detected using quantitative real-time polymerase chain reaction (qRT-PCR). The proliferation, migration, and invasion of infiltrative GC cells (XGC-1) were determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide (MTT), plate clone, wound-healing, or transwell assays. Protein levels were detected by Western blotting. Measurements of extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) were executed using an XF96 extracellular flux analyzer. Glucose uptake and lactate production were analyzed by glycolysis assay. The regulatory mechanism of circVPS33B had been explored by bioinformatics analysis, dual-luciferase reporter assay, and/or RNA pull-down assay. In vivo tumorigenesis assay was executed to verify the oncogenicity of circVPS33B.
Results: CircVPS33B was upregulated in infiltrative GC tissues and cells. CircVPS33B silencing decreased tumor growth in vivo and inhibited proliferation, migration, invasion, EMT, and Warburg effect of infiltrative GC cells in vitro. Mechanically, circVPS33B regulated heterogeneous nuclear ribonucleoprotein K (HNRNPK) expression via sponging miR-873-5p. Furthermore, miR-873-5p inhibitor offset circVPS33B knockdown-mediated effects on malignant behaviors and Warburg effect of infiltrative GC cells. HNRNPK overexpression reversed the inhibitory impact of miR-873-5p mimic on malignant behaviors and Warburg effect of infiltrative GC cells.
Conclusion: CircVPS33B accelerated Warburg effect and tumor growth through regulating the miR-873-5p/HNRNPK axis in infiltrative GC, manifesting that circVPS33B might be a potential target for infiltrative GC treatment.
Keywords: circVPS33B, miR-873-5p, HNRNPK, infiltrative GC
Introduction
Gastric cancer (GC), which is a malignant tumor, ranks third among cancer-related death causes.1 GC is divided into infiltrative and expanding types according to the growth and invasive patterns based on Ming’s classification.2 Expanding GC exhibits a massive expansion growth pattern, usually forming plenty discrete tumor nodules in tumor tissues, while infiltrative GC presents scattered infiltrating growth without obvious aggregation.3 Moreover, the prognosis of infiltrative GC patients is worse than that of expanding GC patients.4 Studies have reported that the recurrence of the peritoneal or liver after radical resection of GC is closely related to the pathological infiltration pattern (INF).5,6 Also, there is an obvious gene regulation mode between expanding GC and infiltrative GC.7
Circular RNAs (circRNAs), endogenous RNA molecules, have closed-loop structures formed by reverse splicing of mRNA exons or introns.8 CircRNAs do not contain 5ʹ or 3ʹ free terminus, so they are resistant to exonuclease and are more stable than other conventional linear RNAs.9 Increasing evidence has indicated that circRNAs exert vital roles in cellular physiology through serving as protein translation templates, transcription regulators, RBP-binding molecules, or microRNA (miR) sponges.10 Researchers have uncovered that the dysregulation of circRNAs is closely related to the advancement of diverse diseases, including cancer.11 For example, circRNA hsa_circ_0003141 contributed to the tumorigenesis of hepatocellular cancer.12 Our previous research revealed that circRNA circHIPK3 was overexpressed in infiltrative GC cells than that in expanding GC cells, and it played a promoting effect on the proliferation of GC cells via downregulating miR-124 or miR-29b.7 CircRNA VPS33B, also termed as hsa_circ_0005529, is derived from the vacuolar protein sorting 33 homologue B (VPS33B) gene (http://www.circbank.cn/search.html?selectValue=hsa_circ_0005529). It was reported that circVPS33B expression was prominently elevated in GC tissues with respect to adjacent noncancerous tissues (GSE78092).13 However, the regulatory mechiansm of circVPS33B in GC progression has not been adequately clucidated.
MiRs usually regulate gene expression post-transcriptionally and act as powerful regulators in a series of cellular processes, including cell development, differentiation, growth, and apoptosis.14,15 Studies have revealed that miRs exert important roles in numerous cancers, including GC.16 MiRs are not only involved in the occurrence of GC but also regulate the growth pattern of GC.17 Chen et al reported that miR-145 expression was significantly lower in infiltrative GC tissues than expanding GC tissues, and miR-145 overexpression repressed GC cell invasion and migration through targeting FSCN1.18 Wang et al uncovered that miR-15a-3p and miR-16-1-3p were lowly expressed in infiltrative GC tissues compared to expanding GC tissues, and miR-15a-3p and miR-16-1-3p overexpression constrained GC cell invasion and migration through targeting Twist1.19 MiR-873-5p has been revealed as an epigenetic modulator in the early stages of liver fibrosis and cirrhosis.20 Also, miR-873-5p plays an inhibitory effect on tumorigenesis in a series of cancers, such as glioma,21 papillary thyroid cancer,22 and colorectal cancer.23 In addition, miR-873-5p acts as a tumor-inhibiting molecular in GC.24,25 However, the regulatory mechanism of miR-873-5p in GC growth pattern is unclear.
Heterogeneous nuclear ribonucleoprotein K (HNRNPK), a DNA/RNA-binding protein, is involved in regulating various biological processes and disease pathogenesis.26 Furthermore, HNRNPK inhibition can elevate DNA damage after radiation, thereby decreasing tumor cell survival.27 Moreover, HNRNPK upregulation is related to the poor prognosis of several cancers, such as bladder cancer28 and colorectal cancer.29 Also, HNRNPK has been revealed as an oncogene in GC.30 Nevertheless, the mechanism of HNRNPK dysregulation in GC development has not been fully elucidated.
In the present study, we verified that circVPS33B expression was observably elevated in infiltrative GC tissues. Mechanically, circVPS33B accelerated tumor growth and Warburg effect via elevating HNRNPK expression through acting as a miR-873-5p sponge. Our findings provided a novel mechanism by which circVPS33B modulated infiltrative GC growth through the miR-873-5p/HNRNPK axis.
Materials and Methods
Patient-Derived Samples
The research was approved by the Ethics Committee of Zhongshan Hospital Xiamen University and executed in accordance with the Declaration of Helsinki. Sixty-three paired GC tissue samples and neighboring non-tumor tissues were collected from the Zhongshan Hospital Xiamen University, including 44 infiltrative GC tissues and 19 expanding GC tissues. These samples were collected from May 2018 to August 2019. All registered patients signed written informed consent and did not receive other treatments before surgery. The clinical features of GC patients are exhibited in Table 1.
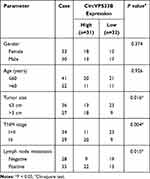 |
Table 1 Association Between Clinical Features and CircVPS33B Expression of GC Patients (n=63) |
Cell Culture and Transfection
Infiltrative GC cells XGC-1 (Patent No.: CN103396994A) was obtained from the Zhongshan Hospital Xiamen University. Human gastric epithelial mucosa cells (GES-1) were purchased from BeNa Culture Collection (Suzhou, China). These cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (HyClone, Logan, UT, USA) supplemented with fetal bovine serum (FBS, 10%, HyClone) and streptomycin/penicillin (1%, Solarbio, Beijing, China) in a moist atmosphere with 5% CO2 at 37°C.
Small interference (si) RNA against circVPS33B (si-circVPS33B) and its negative control (NC) (si-NC), as well as short hairpin (sh) targeting circVPS33B (sh-circVPS33B) and matched NC (sh-NC), were purchased from GenePharma (Shanghai, China). To generate the pcDNA-HNRNPK plasmid, the full-length sequence of HNRNPK was cloned into empty pcDNA3.1 vector (pcDNA-NC) (Life Technologies, Grand Island, NY, USA). MiR-873-5p mimic (miR-873-5p), miR-873-5p inhibitor (anti-miR-873-5p), and their corresponding negative controls (miR-NC and anti-miR-NC) were purchased from RiboBio (Guangzhou, China). Transient transfection was performed using the Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA). XGC-1 cells with stable knockdown of circVPS33B were produced by infection with lentiviral particles, followed by selection with puromycin (Solarbio).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
RNA Simple Total Kit (TIANGEN, Beijing, China) was utilized for total RNA separation (tissue samples and cells). The complementary DNA was synthesized with the M-MLV First Strand Kit (Life Technologies) or Bulge-Loop miR RT-qPCR Starter kit (RiboBio), followed by performing qPCR with the SYBR Green (Solarbio). Primer sequences used in the study were as follows: circVPS33B (Forward: 5ʹ-GTGGTGTTCTTGGGTGGTTGT-3ʹ; Reverse: 5ʹ-CCGCTCTAGCACCTTTCTCTC-3ʹ), VPS33B (Forward: 5ʹ-ATGAGCCCTTTGGATCGAATTG-3ʹ; Reverse: 5ʹ-ATGCGGGGTCTGACCAAGA-3ʹ), HNRNPK (Forward: 5ʹ-CAATGGTGAATTTGGTAAACGCC-3ʹ; Reverse: 5ʹ-GTAGTCTGTACGGAGAGCCTTA-3ʹ), β-actin (Forward: 5ʹ-CTCGCCTTTGCCGATCC-3ʹ; Reverse: 5ʹ-TCTCCATGTCGTCCCAGTTG-3ʹ), miR-873-5p (Forward: 5ʹ-GGGGCAGGAACTTGTGAG-3ʹ; Reverse: 5ʹ-GTGTGGTGTGGTATGGTGTG-3ʹ), and U6 small nuclear RNA (U6) (Forward: 5ʹ-CGCTTCGGCAGCACATATACTAAAATTGGAAC-3ʹ; Reverse: 5ʹ-GCTTCACGAATTTGCGTGTCATCCTTGC-3ʹ). Relative expression of circVPS33B, VPS33B mRNA, HNRNPK mRNA, and miR-873-5p was evaluated by 2−ΔΔCt method, and β-actin or U6 was used as an internal reference.
Actinomycin D Treatment and RNase R Digestion
To verify the stability of circVPS33B in XGC-1 cells, we performed Actinomycin D treatment and RNase R digestion. For actinomycin D treatment, XGC-1 cells were cultured in a complete medium supplemented with 2 μg/mL actinomycin D (Sigma, St Louis, MO, USA) or dimethyl sulfoxide (Sigma). For RNA digestion, total RNA from XGC-1 cells was incubated with 3 U/μg RNase R (Geneseed, Guangzhou, China) or diethylpyrocarbonate-treated water (Sigma).
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazoliumbromide (MTT) Assay
XGC-1 cells (5×103 cells) were seeded into 96-well plates and cultured for 24 h, 48 h, or 72 h. After discarding the supernatant, the MTT solution (100 μL, 0.5 mg/mL, Sigma) was added to each well and incubated for 4 h. Then, the purple crystals were dissolved with dimethyl sulfoxide (150 μL, Sigma). Next, the absorbance at 570 nm was evaluated by the Microplate Reader (Bio-Rad, Hercules, CA, USA).
Plate Clone Assay
XGC-1 cells (5×103 cells/well) were seeded onto 6-well plates. Two weeks later, XGC-1 cells were fixed with paraformaldehyde (4%, Sigma) and then stained with crystal violet (0.5%, Sigma). The number of colonies was counted with an inverted microscope (Nikon, Tokyo, Japan).
Wound-Healing and Transwell Assays
The migration of XGC-1 cells was analyzed by wound-healing and transwell assays. For the wound-healing assay, wounds on cell layer were created using a pipette tip (100 μL) when the cells reached 90% confluence. Then, FBS-free medium was used to rinse the created wound to remove all exfoliated cell debris. Photographs were taken at 0 h and 24 h with an inverted microscope (Nikon). Percentage of migration was calculated according to the following equation: cell migration rate (%) = (1-the distance following healing/the distance prior to healing) × 100%.
For the transwell migration assay, serum-free medium containing transfected XGC-1 cells (1×105 cells) was added to the top chamber (8 μm, Costar, Cambridge, MA, USA). The cell medium containing 10% FBS was added to the bottom chamber. After removing the cells on the upper surface of the membrane, the remaining cells were fixed with paraformaldehyde (4%, Sigma) and stained with crystal violet (0.5%, Sigma). The number of migrating cells was calculated with an inverted microscope (Nikon) at 100 × magnification.
The procedure of the transwell invasion assay was the same as the transwell migration assay. It was worth noting that the transwell chamber used in the invasion assay had been pre-coated with Matrigel (Sigma).
Western Blotting
Total protein was separated using the RIPA lysis buffer (Solarbio). All antibodies utilized in the research were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA), including anti-β-actin (sc-8432), anti-ki-67 (sc-23900), anti-E-cadherin (sc-21791), anti-N-cadherin (sc-59987), anti-vimentin (sc-373717), and anti-HNRNPK (sc-28380). Also, m-IgG BP-HRP (sc-516102) was used as a secondary antibody. Western blotting was executed as previously described.31 The immunoblot was visualized through the enhanced chemiluminescence reagent kit (Beyotime, Shanghai, China).
Analysis of Extracellular Acidification Rate (ECAR) and Oxygen Consumption Rate (OCR)
The ECAR and OCR of transfected XGC-1 cells were analyzed using the Seahorse XF Glycolysis Stress Test Kit (Seahorse Bioscience, Chicopee, MA, USA) or Seahorse XF Cell Mito Stress Test Kit (Seahorse Bioscience) on an XF96 Extracellular Flux analyzer (Seahorse Bioscience). In short, the cells (1×104) were seeded into Seahorse XF 96 cell culture microplates. For the ECAR analysis, glucose, oligomycin, and 2-deoxy-D-glucose (2-DG) were sequentially injected into each well at the specified time point. For the OCR analysis, oligomycin, p-trifluoromethoxy carbonyl cyanide phenylhydrazone (FCCP), and rotenone (Rote) plus antimycin A (AA) were sequentially injected into each well after baseline measurement. At last, the data were analyzed with the Seahorse XF-96 Wave software, and ECAR and OCR were presented in mpH/min and pmoles/min, respectively.
Glycolysis Assay
The supernatant of transfected XGC-1 cells was collected and the concentrations of glucose and lactate in the supernatant were assessed with a Glucose Assay kit (Sigma) or Lactate Assay kit (BioVision, Mountain View, CA, USA) according to the manufacturer’s instructions. Glucose uptake was the difference between the glucose concentration in the medium and the control group.
Subcellular Fractionation and Localization
The subcellular fractionation of XGC-1 cells was performed with the PARIS kit (Life Technologies) based on the manufacturer’s instructions. Expression of circVPS33B in the nuclear and cytoplasm of XGC-1 cells was detected by qRT-PCR, and U6 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control for nuclear and cytoplasm. The primer sequences for GAPDH were as follows: (Forward: 5ʹ-GATTCCACCCATGGCAAATTCC-3ʹ; Reverse: 5ʹ-TCGCTCCTGGAAGATGGTGAT-3ʹ)
Dual-Luciferase Reporter Assay
The binding sites of circVPS33B in miR-873-5p were predicted with the circInteractome database. The binding sites between HNRNPK and miR-873-5p were jointly predicted with the TarBase, TargetScan, miRDB, and miRWalk databases. The fragments of wild type (WT) circVPS33B (circVPS33B-WT), mutant (MUT) circVPS33B (circVPS33B-MUT), HNRNPK 3ʹUTR-WT, and HNRNPK 3ʹUTR-MUT were synthesized and inserted into the psiCHECK-2 vectors (Promega, Madison, WI, USA), respectively. The luciferase activity in the lysate of XGC-1 cells co-transfected with a luciferase plasmid and miR-873-5p mimic or miR-NC was evaluated by using the luciferase reporter assay kit (Promega).
RNA Pull-Down Assay
The biotinylated (Bio)-miR-873-5p, Bio-miR-NC, and Bio-miR-873-5p-MUT probes were bought from Sigma. The lysates of XGC-1 cells were incubated with probe-coated beads generated by incubating a probe with the M-280 Streptavidin magnetic beads (Invitrogen). The RNA complexes were isolated with TRIzol reagent (Solarbio), and the abundance of circVPS33B in RNA complexes was assessed by qRT-PCR analysis.
In vivo Tumorigenesis Assay
The protocols of xenograft assay were authorized by the Animal Ethics Committee of the Zhongshan Hospital Xiamen University and executed in accordance with the Laboratory-animal-Guideline for ethical review of animal welfare (GB/T 35892–2018). Ten BALB/c nude mice (4-week-old) were bought from Vital River Laboratory Animal Co., Ltd. (Beijing, China) and randomly divided into two groups. For the xenograft assay, XGC-1 cells (1×106) with sh-circVPS33B or sh-NC were subcutaneously injected into the right flank of BALB/c nude mice. Tumor volume was measured once a week with a caliper from the second week after injection. Five weeks later, all mice were euthanized by cervical dislocation under isoflurane (5%) treatment and then taken their tumor tissues. IHC was performed as previously described with the antibody against HNRNPK (sc-28380).32 Tumors volume was counted based on the following equation: Volume = (length × width2)/2.
Statistical Analysis
Statistical analysis was implemented with GraphPad Prism 6 software (GraphPad, San Diego, CA, USA). The experiments in vitro were repeated at least 3 times and each experiment was conducted in triplicate. Data were exhibited as the mean ± standard deviation. Correlation among circVPS33B, HNRNPK, and miR-873-5p in infiltrative GC tissues were evaluated with the Pearson’s correlation analysis. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test was utilized to assess the differences among 3 or more groups. Unpaired Student’s t-test was applied to compare the difference between 2 groups. Association between clinical features and circVPS33B expression of infiltrative GC patients was analyzed by Chi-square test. Difference was deemed significant if P < 0.05.
Results
High circVPS33B Expression Might Be Associated with the Development of Infiltrative GC
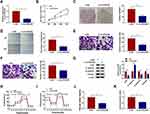
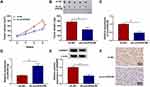
Through searching the public database (GES78092), we noted that there were 199 differentially expression circRNAs in GC tissues (Figure 1A). Among these differentially expression circRNAs, 20 differentially expressed circRNAs were picked out to draw a heat map, as exhibited in Figure 1B. CircVPS33B was a significantly differentially expressed circRNA, so it was selected as a candidate gene for subsequent analysis. CircVPS33B (193 bp), located at chr15: 91542906–91543205, is derived from the VPS33B gene (Figure 1C). To validate the expression pattern of circVPS33B in GC, we performed qRT-PCR analysis to detect circVPS33B expression in 63 paired tissue samples The results exhibited that circVPS33B expression was significantly elevated in GC tissues (infiltrative GC tissues and expanding GC tissues) in contrast to the neighboring non-tumor tissues, and there was an observable upregulation in circVPS33B expression in infiltrative GC tissues with respect to expanding GC tissues (Figure 1D). Furthermore, high circVPS33B expression was associated with tumor size, TNM stage, and lymph node metastasis of infiltrative GC patients (Table 1). Similarly, circVPS33B expression was overtly increased in XGC-1 cells compared to the GES-1 cells (Figure 1E). Actinomycin D assay revealed that circVPS33B had a longer half-time than linear VPS33B in XGC-1 cells (Figure 1F). RNase R digestion assay presented that linear VPS33B was degraded after RNase R treatment, whereas circVPS33B was resistant to RNase R (Figure 1G). These data indicated that high circVPS33B expression might be associated with the development of infiltrative GC.
Inhibition of circVPS33B Reduced the Malignancy and Warburg Effect of Infiltrative GC Cells
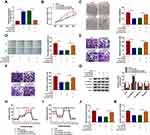
Subsequently, we investigated the role of circVPS33B in infiltrative GC via loss-of-function assays. The knockdown efficiency of si-circVPS33B was verified by qRT-PCR and the results displayed that circVPS33B expression was prominently reduced in XGC-1 cells after si-circVPS33B transfection compared with the control group (Figure 2A). MTT and plate clone assays exhibited that circVPS33B downregulation inhibited the proliferation of XGC-1 cells (Figure 2B and C). Wound-healing and transwell assays showed that the migration and invasion of XGC-1 cells was repressed after circVPS33B knockdown (Figure 2D–F). EMT, the process by which epithelial cells acquire mesenchymal characteristics, is related to tumor initiation, invasion, and metastasis.33 N-cadherin and Vimentin are usually upregulated during EMT, while E-cadherin is usually downregulated.34 Ki-67 is an excellent marker for determining cell growth.35 Results of Western blotting showed that the protein levels of ki-67, N-cadherin, and Vimentin were downregulated in circVPS33B-silenced XGC-1 cells, while the protein level of E-cadherin was upregulated (Figure 2G). Moreover, we also analyzed the influence of circVPS33B inhibition on the Warburg effect of infiltrative GC cells. The results exhibited that circVPS33B knockdown resulted in a decrease in ECAR and an increase in OCR in XGC-1 cells (Figure 2H and I). Furthermore, circVPS33B silencing reduced glucose uptake and lactate production in XGC-1 cells (Figure 2J and K). Taken together, these data indicated that circVPS33B knockdown repressed proliferation, migration, invasion, EMT, and decreased Warburg effect of infiltrative GC cells.
CircVPS33B Was Identified as a miR-873-5p Sponge
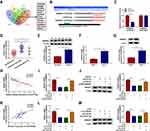
In consideration of the above findings, we further explored the regulatory mechanism of circVPS33B in infiltrative GC. Subcellular fractionation and qRT-PCR presented that circVPS33B was mainly distributed in the cytoplasm of XGC-1 cells, indicating that circVPS33B might play its function by acting as a miR sponge (Figure 3A). Through circInteractome prediction, we discovered that miR-873-5p might interact with circVPS33B (Figure 3B). In addition, miR-873-5p was markedly upregulated after transfection with miR-873-5p mimic in XGC-1 cells (Figure 3C). Dual-luciferase reporter assay indicated that miR-873-5p overexpression repressed the luciferase intensity of the luciferase plasmid containing circVPS33B-WT in XGC-1 cells, whereas the luciferase intensity of the luciferase plasmid with circVPS33B-MUT did not change (Figure 3D). RNA pull-down assay presented that circVPS33B could be pulled down by Bio-miR-873-5p probe instead of Bio-miR-NC and Bio-miR-873-5p-MUT probes (Figure 3E). Moreover, there was a marked downregulation in miR-873-5p expression in GC tissues (GES78091) (Figure 3F). As expected, miR-873-5p expression was overtly decreased in infiltrative GC tissues compared to expanding GC tissues (Figure 3G). Similarly, miR-873-5p was downregulated in XGC-1 cells than that in GES-1 cells (Figure 3H). Pearson’s correlation analysis indicated that miR-873-5p and circVPS33B had a negative correlation in infiltrative GC tissues (Figure 3I). Furthermore, circVPS33B inhibition elevated the expression of miR-873-5p in XGC-1 cells (Figure 3J). However, miR-873-5p mimic had no effect on circVPS33B expression in XGC-1 cells (Figure 3K). Collectively, these findings manifested that circVPS33B acted as a sponge for miR-873-5p in infiltrative GC cells.
CircVPS33B Facilitated the Malignancy and Warburg Effect of Infiltrative GC Cells by Sponging miR-873-5p
Given that circVPS33B acted as a sponge for miR-873-5p in XGC-1 cells, we further explored whether circVPS33B regulated the malignancy and Warburg effect of infiltrative GC cells via binding to miR-873-5p. We discovered that miR-873-5p inhibitor reversed the upregulation of miR-873-5p in XGC-1 cells caused by circVPS33B knockdown (Figure 4A). Moreover, knockdown of miR-873-5p overturned the inhibiting impact of circVPS33B silencing on proliferation, migration, and invasion of XGC-1 cells (Figure 4B–F). Also, the upregulation of E-cadherin and the downregulation of ki-67, N-cadherin, and Vimentin in XGC-1 cells caused by circVPS33B inhibition were restored after miR-873-5p knockdown (Figure 4G). Furthermore, miR-873-5p inhibitor abolished circVPS33B silencing-mediated effects on ECAR and OCR of XGC-1 cells (Figure 4H and I). Also, the repressive effect of circVPS33B knockdown on glucose uptake and lactate production of XGC-1 cells was overturned by miR-873-5p downregulation (Figure 4J and K). Overall, these findings manifested that circVPS33B modulated the malignancy and Warburg effect of infiltrative GC cells via sponging miR-873-5p.
CircVPS33B Regulated HNRNPK Expression Through Adsorbing miR-873-5p
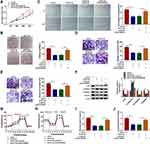
Next, the downstream targets of miR-873-5p were jointly predicted by bioinformatics analysis (TarBase, TargetScan, miRDB, and miRWalk). As shown in Figure 5A, 13 genes might be downstream targets of miR-873-5p. Based on the pre-experiment, we chose HNRNPK for further research. The complementary-binding sites between miR-873-5p and HNRNPK are displayed in Figure 5B. Moreover, the luciferase activity of the luciferase plasmid containing HNRNPK 3ʹUTR-WT was inhibited in XGC-1 cells in the presence of miR-873-5p mimic, but there was no overt difference in the luciferase plasmid containing HNRNPK 3ʹUTR-MUT (Figure 5C). We also observed that the expression of HNRNPK mRNA was elevated in infiltrative GC tissues compared to expanding GC tissues (Figure 5D). There was also a significant upregulation in HNRNPK at protein level in GC tissues (Figure 5E). Also, the levels of HNRNPK mRNA and protein were increased in XGC-1 cells than that in GES-1 cells (Figure 5F and G). As expected, the expression of HNRNPK mRNA and miR-873-5p had a negative correlation in infiltrative GC tissues (Figure 5H). Furthermore, miR-873-5p mimic reduced the levels of HNRNPK mRNA and protein in XGC-1 cells, while this decrease was reversed after pcDNA-HNRNPK transfection (Figure 5I and J). In addition, there was a positive correlation between circVPS33B and HNRNPK mRNA in infiltrative GC tissues (Figure 5K). And miR-873-5p inhibitor overturned the downregulation of HNRNPK in circVPS33B-silenced XGC-1 cells (Figure 5L and M). Collectively, these results manifested that circVPS33B regulated HNRNPK expression via sponging miR-873-5p in infiltrative GC cells.
MiR-873-5p Constrained the Malignancy and Warburg Effect of Infiltrative GC Cells by Targeting HNRNPK
Based on the above findings, we further investigated whether miR-873-5p regulated the malignancy and Warburg effect of XGC-1 cells via targeting HNRNPK. The results exhibited that the repressive impact of miR-873-5p mimic on cell proliferation, migration, and invasion in XGC-1 cells was reversed by HNRNPK overexpression (Figure 6A–E). Moreover, miR-873-5p overexpression reduced protein levels of ki-67, N-cadherin, Vimentin, and increased protein level of E-cadherin in XGC-1 cells, but these impacts caused by miR-873-5p mimic were overturned after HNRNPK overexpression (Figure 6F). Additionally, miR-873-5p mimic decreased ECAR and elevated OCR in XGC-1 cells, but these tendencies were abolished by HNRNPK overexpression (Figure 6G and H). Furthermore, overexpression of HNRNPK restored the decrease of glucose uptake and lactate production in XGC-1 cells mediated by miR-873-5p upregulation (Figure 6I and J). Overall, these data indicated that miR-873-5p regulated the malignancy and Warburg effect of infiltrative GC cells via targeting HNRNPK.
Knockdown of circVPS33B Reduced Infiltrative GC Growth in vivo
To verify the oncogenicity of circVPS33B, we injected XGC-1 cells that stably knockdown sh-circVPS33B or sh-NC into BALB/c nude mice. The results presented that tumor volume and weight were prominently smaller and lighter in the sh-circVPS33B group in contrast to the sh-NC group (Figure 7A and B). Also, the expression of circVPS33B was overtly downregulated in mouse tumor tissues in the sh-circVPS33B group compared with the control group, but the expression of miR-873-5p had an opposite tendency (Figure 7C and D). Western blotting and IHC exhibited that HNRNPK was also observably reduced in mouse tumor tissues in the sh-circVPS33B group with respect to the control group (Figure 7E and F). Together, these data manifested that circVPS33B inhibition decreased infiltrative GC growth in vivo.
Discussion
In the current work, we proved that circVPS33B acted as a miR-873-5p sponge and elevated HNRNPK expression via adsorbing miR-873-5p, thus accelerating Warburg effect and tumor growth of infiltrative GC.
Warburg effect, also known as aerobic glycolysis, exhibits extremely high glucose uptake and lactate fermentation in an aerobic environment.36 The Warburg effect provides energy and a favorable environment for the continuous proliferation and malignant progression of cancer cells.37 The Warburg effect is usually accompanied by a decrease in OCR and an increase in ECAR in cancer cells.38 A recent study reported that circVPS33B expression was significantly elevated in GC tissues, and circVPS33B elevated Sp1 expression via adsorbing miR-527, thus promoted cancer cell growth and migration.14,39 Herein, circVPS33B was also upregulated in GC tissues, and we also observed that circVPS33B expression was significantly higher in infiltrative GC tissues than expanding GC tissues. Also, high circVPS33B expression was associated with tumor size, TNM stage, and lymph node metastasis of infiltrative GC patients, manifesting that circVPS33B upregulation might be related to the advancement of infiltrative GC. Furthermore, circVPS33B knockdown decreased infiltrative GC cell growth in vivo and repressed infiltrative GC cell proliferation, migration, invasion, and EMT in vitro. Also, circVPS33B silencing decreased ECAR, glucose uptake, lactate production in infiltrative GC cells, while increased OCR, indicating that circVPS33B knockdown decreased the Warburg effect of infiltrative GC cells. Therefore, we concluded that circVPS33B played a promoting impact on the malignancy and Warburg effect of infiltrative GC.
Accumulated evidence suggests that circRNAs can regulate gene expression by serving as miR sponges.12,40,41 In this study, circVPS33B was preferentially located in the cytoplasm of infiltrative GC cells, suggesting that circVPS33B might act as a miR sponge. Through bioinformatics analysis, dual-luciferase reporter assay, and RNA pull down assay, we verified that circVPS33B was a miR-873-5p sponge. MiR-873-5p has been proved to exert a tumor-inhibiting role in a variety of cancers. Zhu et al indicated that miR-873-5p curbed colon cancer progression via repressing the TUSC3/AKT pathway.42 Also, miR-873-5p targeted CXCL16 to impede the progression of papillary thyroid cancer.22 Furthermore, lncRNA TDRG1 and lncRNA DDX11-AS1 accelerated the aggressiveness of GC by sponging miR-873-5p.24,25 Herein, miR-873-5p was lowly expressed in infiltrative GC tissues. Downregulation of miR-873-5p partly neutralized the repressive impact of circVPS33B silencing on the malignancy and Warburg effect of infiltrative GC cells. Therefore, we inferred that circVPS33B sponged miR-873-5p to modulate the malignancy and Warburg effect of infiltrative GC.
Subsequently, we found that HNRNPK was a miR-873-5p target. HNRNPK had been proved to be associated with tumor stage and metastasis in GC. Moreover, HNRNPK elevation contributed to invasion, migration, and proliferation of GC cells via regulating the alternative splicing of CD44E.30 Also, HNRNPK affected mitochondrial function and tumor progression through splicing MRPL33.43 Our data presented that HNRNPK was highly expressed in infiltrative GC tissues in comparison to expanding GC tissues. Moreover, overexpression of HNRNPK reversed the inhibitory effect of miR-873-5p mimic on the malignancy and Warburg effect of infiltrative GC cells. Also, circVPS33B modulated HNRNPK expression via competitively binding to miR-873-5p. Overall, we concluded that circVPS33B modulated tumor growth and Warburg effect via sponging miR-873-5p and elevating HNRNPK expression in infiltrative GC.
Conclusion
In sum, circVPS33B played a promoting impact on infiltrative GC growth. Mechanically, circVPS33B adsorbed miR-873-5p to increase HNRNPK expression, thereby accelerating tumor growth and Warburg effect of infiltrative GC. The research indicated that circVPS33B might be a hopeful target for infiltrative GC treatment. Additionally, whether circVPS33B regulated the malignancy and Warburg effect of infiltrative GC through exosomes and HNRNPK-mediated CD44E and MRPL33 could be further explored in the future.
Data Sharing Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The present study was approved by the Ethics Committee of the Zhongshan Hospital Xiamen University. Written informed consent was obtained from all enrolled patients.
Funding
This work was supported by grant from The National Natural Science Foundation of China (Nos. 81172283 and 81871979); the Natural Science Foundation of Fujian Province (no. 2020J011215); Fujian Provincial Medical Innovation Project (No. 2017-CXB-15); Guiding Project of Xiamen Science and Technology Plan (3502Z20149005 3502Z20174076); Young and Middle-aged Backbone Key Research Project of National Health and Family Planning Commission of Fujian Province (2017-ZQN-89).
Disclosure
The authors declare that they have no competing interests.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Piard F, Hillon P, Levillain P, et al. [Does Ming’s classification of gastric carcinomas have epidemiologic or prognostic value?]. Ann Pathol. 1986;6(4–5):329–334.
3. Ming SC. Gastric carcinoma. A pathobiological classification. Cancer. 1977;39(6):2475–2485. doi:10.1002/1097-0142(197706)39:6<2475::AID-CNCR2820390626>3.0.CO;2-L
4. Luebke T, Baldus SE, Grass G, et al. Histological grading in gastric cancer by Ming classification: correlation with histopathological subtypes, metastasis, and prognosis. World J Surg. 2005;29(11):1422–1427. doi:10.1007/s00268-005-7795-z
5. Kanda M, Mizuno A, Fujii T, et al. Tumor infiltrative pattern predicts sites of recurrence after curative gastrectomy for stages 2 and 3 gastric cancer. Ann Surg Oncol. 2016;23(6):1934–1940. doi:10.1245/s10434-016-5102-x
6. Nakagawa N, Kanda M, Ito S, et al. Pathological tumor infiltrative pattern and sites of initial recurrence in stage II/III gastric cancer: propensity score matching analysis of a multi-institutional dataset. Cancer Med. 2018;7(12):6020–6029. doi:10.1002/cam4.1868
7. Cheng J, Zhuo H, Xu M, et al. Regulatory network of circRNA-miRNA-mRNA contributes to the histological classification and disease progression in gastric cancer. J Transl Med. 2018;16(1):216. doi:10.1186/s12967-018-1582-8
8. Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi:10.1016/j.molcel.2018.06.034
9. Li HM, Ma XL, Li HG. Intriguing circles: conflicts and controversies in circular RNA research. Wiley Interdiscip Rev RNA. 2019;10(5):e1538. doi:10.1002/wrna.1538
10. Zhao W, Dong M, Pan J, et al. Circular RNAs: a novel target among non‑coding RNAs with potential roles in malignant tumors (Review). Mol Med Rep. 2019;20(4):3463–3474. doi:10.3892/mmr.2019.10637
11. Wang Y, Liu J, Ma J, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18(1):116. doi:10.1186/s12943-019-1041-z
12. Wang Y, Gao R, Li J, et al. Circular RNA hsa_circ_0003141 promotes tumorigenesis of hepatocellular carcinoma via a miR-1827/UBAP2 axis. Aging (Albany NY). 2020;12:9793.
13. Huang YS, Jie N, Zou KJ, Weng Y. Expression profile of circular RNAs in human gastric cancer tissues. Mol Med Rep. 2017;16(3):2469–2476. doi:10.3892/mmr.2017.6916
14. Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234(5):5451–5465. doi:10.1002/jcp.27486
15. Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202–1207. doi:10.1016/j.jaci.2017.08.034
16. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi:10.1038/nrd.2016.246
17. Cheng J, Zhuo H, Wang L, et al. Identification of the combinatorial effect of miRNA Family regulatory network in different growth patterns of GC. Mol Ther Oncolytics. 2020;17:531–546. doi:10.1016/j.omto.2020.03.012
18. Chen J, Cai W, Liu X, et al. Reverse correlation between microRNA-145 and FSCN1 affecting gastric cancer migration and invasion. PLoS One. 2015;10(5):e0126890. doi:10.1371/journal.pone.0126890
19. Wang T, Hou J, Li Z, et al. miR-15a-3p and miR-16-1-3p negatively regulate twist1 to repress gastric cancer cell invasion and metastasis. Int J Biol Sci. 2017;13(1):122–134. doi:10.7150/ijbs.14770
20. Fernández-Ramos D, Fernández-Tussy P, Lopitz-Otsoa F, et al. MiR-873-5p acts as an epigenetic regulator in early stages of liver fibrosis and cirrhosis. Cell Death Dis. 2018;9(10):958. doi:10.1038/s41419-018-1014-y
21. Lin YH, Guo L, Yan F, Dou ZQ, Yu Q, Chen G. Long non-coding RNA HOTAIRM1 promotes proliferation and inhibits apoptosis of glioma cells by regulating the miR-873-5p/ZEB2 axis. Chin Med J. 2020;133(2):174–182. doi:10.1097/CM9.0000000000000615
22. Wang Z, Liu W, Wang C, Ai Z. miR-873-5p inhibits cell migration and invasion of papillary thyroid cancer via regulation of CXCL16. Onco Targets Ther. 2020;13:1037–1046. doi:10.2147/OTT.S213168
23. Wang L, Jiang F, Ma F, Zhang B. MiR-873-5p suppresses cell proliferation and epithelial-mesenchymal transition via directly targeting Jumonji domain-containing protein 8 through the NF-κB pathway in colorectal cancer. J Cell Commun Signal. 2019;13(4):549–560. doi:10.1007/s12079-019-00522-w
24. Ma Y, Xu XL, Huang HG, Li YF, Li ZG. LncRNA TDRG1 promotes the aggressiveness of gastric carcinoma through regulating miR-873-5p/HDGF axis. Biomed Pharmacother. 2020;121:109425. doi:10.1016/j.biopha.2019.109425
25. Ren Z, Liu X, Si Y, Yang D. Long non-coding RNA DDX11-AS1 facilitates gastric cancer progression by regulating miR-873-5p/SPC18 axis. Artif Cells Nanomed Biotechnol. 2020;48(1):572–583. doi:10.1080/21691401.2020.1726937
26. Wang Z, Qiu H, He J, et al. The emerging roles of hnRNPK. J Cell Physiol. 2020;235(3):1995–2008. doi:10.1002/jcp.29186
27. Wiesmann N, Strozynski J, Beck C, et al. Knockdown of hnRNPK leads to increased DNA damage after irradiation and reduces survival of tumor cells. Carcinogenesis. 2017;38(3):321–328. doi:10.1093/carcin/bgx006
28. Chen X, Gu P, Xie R, et al. Heterogeneous nuclear ribonucleoprotein K is associated with poor prognosis and regulates proliferation and apoptosis in bladder cancer. J Cell Mol Med. 2017;21(7):1266–1279. doi:10.1111/jcmm.12999
29. Gao T, Liu X, He B, et al. Exosomal lncRNA 91H is associated with poor development in colorectal cancer by modifying HNRNPK expression. Cancer Cell Int. 2018;18:11. doi:10.1186/s12935-018-0506-2
30. Peng WZ, Liu JX, Li CF, Ma R, Jie JZ. hnRNPK promotes gastric tumorigenesis through regulating CD44E alternative splicing. Cancer Cell Int. 2019;19:335. doi:10.1186/s12935-019-1020-x
31. Zhou M, Zhang XY, Yu X. Overexpression of the long non-coding RNA SPRY4-IT1 promotes tumor cell proliferation and invasion by activating EZH2 in hepatocellular carcinoma. Biomed Pharmacother. 2017;85:348–354. doi:10.1016/j.biopha.2016.11.035
32. Seung BJ, Lim HY, Shin JI, et al. CD204-expressing tumor-associated macrophages are associated with malignant, high-grade, and hormone receptor-negative canine mammary gland tumors. Vet Pathol. 2018;55(3):417–424. doi:10.1177/0300985817750457
33. Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29(3):212–226. doi:10.1016/j.tcb.2018.12.001
34. Maeda M, Johnson KR, Wheelock MJ. Cadherin switching: essential for behavioral but not morphological changes during an epithelium-to-mesenchyme transition. J Cell Sci. 2005;118(5):873–887. doi:10.1242/jcs.01634
35. Menon SS, Guruvayoorappan C, Sakthivel KM, Rasmi RR. Ki-67 protein as a tumour proliferation marker. Clin Chim Acta. 2019;491:39–45. doi:10.1016/j.cca.2019.01.011
36. Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41(3):211–218. doi:10.1016/j.tibs.2015.12.001
37. Vaupel P, Schmidberger H, Mayer A. The Warburg effect: essential part of metabolic reprogramming and central contributor to cancer progression. Int J Radiat Biol. 2019;95(7):912–919. doi:10.1080/09553002.2019.1589653
38. Nimmakayala RK, Leon F, Rachagani S, et al. Metabolic programming of distinct cancer stem cells promotes metastasis of pancreatic ductal adenocarcinoma. Oncogene. 2021;40(1):215–231. doi:10.1038/s41388-020-01518-2
39. Zhang X, Yang H, Jia Y, et al. circRNA_0005529 facilitates growth and metastasis of gastric cancer via regulating miR-527/Sp1 axis. BMC Mol Cell Biol. 2021;22(1);6: Published 2021 Jan 20. doi:10.1186/s12860-020-00340-8
40. Liu J, Liu H, Zeng Q, Xu P, Liu M, Yang N. Circular RNA circ-MAT2B facilitates glycolysis and growth of gastric cancer through regulating the miR-515-5p/HIF-1α axis. Cancer Cell Int. 2020;20:171. doi:10.1186/s12935-020-01256-1
41. Luo Z, Rong Z, Zhang J, et al. Circular RNA circCCDC9 acts as a miR-6792-3p sponge to suppress the progression of gastric cancer through regulating CAV1 expression. Mol Cancer. 2020;19(1):86. doi:10.1186/s12943-020-01203-8
42. Zhu Y, Zhang X, Qi M, Zhang Y, Ding F. miR-873-5p inhibits the progression of colon cancer via repression of tumor suppressor candidate 3/AKT signaling. J Gastroenterol Hepatol. 2019;34(12):2126–2134. doi:10.1111/jgh.14697
43. Liu L, Luo C, Luo Y, et al. MRPL33 and its splicing regulator hnRNPK are required for mitochondria function and implicated in tumor progression. Oncogene. 2018;37(1):86–94. doi:10.1038/onc.2017.314
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.