Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 12
Individualized Drug Repositioning For Rheumatoid Arthritis Using Weighted Kolmogorov–Smirnov Algorithm
Authors Hu RY, Tian XB, Li B, Luo R, Zhang B, Zhao JM
Received 11 September 2019
Accepted for publication 6 November 2019
Published 11 December 2019 Volume 2019:12 Pages 369—375
DOI https://doi.org/10.2147/PGPM.S230751
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Ru-Yin Hu,1–3,* Xiao-Bin Tian,3,* Bo Li,3 Rui Luo,3 Bin Zhang,3 Jin-Min Zhao1
1Department of Orthopaedics, Guangxi Medical University, Nanning 530021, People’s Republic of China; 2Department of Orthopaedics, The First Affiliated Hospital of Guangxi Medical University, Nanning 530021, People’s Republic of China; 3Department of Orthopaedics, Guizhou Provincial People’s Hospital, Guiyang 550002, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jin-Min Zhao
Department of Orthopaedics, Guangxi Medical University, No. 22 Shuangyong Road, Nanning, Guangxi 530021, People’s Republic of China
Tel +86 771 13985048001
Email [email protected]
Background: Existing drugs are far from enough for investigators and patients to administrate the therapy of rheumatoid arthritis. Drug repositioning has drawn broad attention by reusing marketed drugs and clinical candidates for new uses.
Purpose: This study attempted to predict candidate drugs for rheumatoid arthritis treatment by mining the similarities of pathway aberrance induced by disease and various drugs, on a personalized or customized basis.
Methods: We firstly measured the individualized pathway aberrance induced by rheumatoid arthritis based on the microarray data and various drugs from CMap database, respectively. Then, the similarities of pathway aberrances between RA and various drugs were calculated using a Kolmogorov–Smirnov weighted enrichment score algorithm.
Results: Using this method, we identified 4 crucial pathways involved in rheumatoid arthritis development and predicted 9 underlying candidate drugs for rheumatoid arthritis treatment. Some candidates with current indications to treat other diseases might be repurposed to treat rheumatoid arthritis and complement the drug group for rheumatoid arthritis.
Conclusion: This study predicts candidate drugs for rheumatoid arthritis treatment through mining the similarities of pathway aberrance induced by disease and various drugs, on a personalized or customized basis. Our framework will provide novel insights in personalized drug discovery for rheumatoid arthritis and contribute to the future application of custom therapeutic decisions.
Keywords: rheumatoid arthritis, drug repositioning, individualized pathway aberrance, differential pathway
Introduction
Rheumatoid arthritis (RA) is a chronic autoimmune disease characterized by joint synovial tissue inflammation associated with the disability of affected joints.1 Patients with RA have an increased mortality, and the expected survival of RA patients is likely to decrease 3–10 years.2 Early diagnosis and effective therapy are critical to prevent joint deterioration and unfavorable disease outcome. Currently, the treatment of RA primarily rests on the use of disease-modifying antirheumatic drugs, and has improved outcomes in RA patients significantly.3–5 Despite significant therapeutic advances in improving the lives of RA patients, RA remains a hard clinical problem because of the accumulated and persistent disease.1 The administration of RA patients needs new drugs for preventative or curative therapies.
Presently, drug repositioning has drawn broad attention from the pharmaceutical companies and research institutes. Relative to the traditional drug development process, drug repositioning replenishes the drying out drug pipelines by reusing marketed drugs and clinical candidates for new uses, such as treating another disease.6 These repositioned drugs with known bioavailability, safety profiles and well-characterized pharmacology can enter clinical trials for alternative indications more rapidly and less risk.7 Currently, multiple computational approaches have been established for drug repositioning.8–10 Connectivity map (CMap) has been widely used in drug repositioning by measuring the similarity in gene expression profiles between compounds in mammalian cell lines.11 While current methods mainly focus on discovering drug candidates targeting huge populations,12,13 personalized therapeutic decisions are scarce.
Human genetics provides insight into disease pathogenesis and guides drug discovery for complex traits.14,15 A large body of evidence points out the influence of inherited genetic factors on both susceptibility and resistance to the disease.14–16 Currently, high-throughput genome-wide association studies have resulted in a paradigm shift in the way that researchers treat complex diseases. Several lines of evidence has revealed numerous genes influencing the likelihood of developing RA by genome-wide analysis.17,18 While most of the biological functionality of the cell arises from complex interactions among genes, and interpreting the consequences on a pathway level has more powerful in understanding how gene activity perturbations account for disease.19,20 Moreover, personalized pathway analysis has been proposed to perform personalized or customized interpretation of disease data,21 making it possible to develop personalized therapeutic decisions.
Here, we attempted to predict candidate drugs for RA treatment from CMap database by mining the similarities of pathway aberrance induced by disease and various drugs, on a personalized or customized basis. Our study will provide novel insights into personalized drug discovery for RA.
Materials And Methods
Data Retrieve
Transcriptome Data Of RA
Here we retrieved the transcriptome data of RA from ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/), under the accession number of E-GEOD-15573.17 In the study of Teixeira et al,17 a complete genome-wide transcript profiling of peripheral blood mononuclear cells from 18 RA patients and 15 controls was conducted using the Illumina Human-6v2 Expression BeadChips. The raw data and the annotations were obtained from the manufacturer’s documents, and the probes were re-annotated to genes symbols.
CMap Data
The CMap is a collection of genome-wide transcriptome data from cultured human cells treated with bioactive small molecules and simple pattern-matching algorithms that discover connections between drugs, genes expression changes and disease phenotypes.11 The CMap contains more than 7000 gene expression profiles for 1309 compounds. All raw data and the annotations were downloaded from the CMap and the expression values for all samples were calculated by affy package R22 with MAS 5.0 normalization. The probes were re-annotated to genes symbols using Brainarray CDF packages.23 Finally, the samples corresponding to the same drugs were merged, and the gene-drug matrix was obtained for subsequent analysis.
Pathway Data
Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/) is a knowledge database for systemic analysis of gene functional information.24 All 300 human pathways (covering 6919 genes) were downloaded from KEGG database.
In the present study, we retrieved transcriptome data of RA, CMap data and KEGG pathway data. Prior to analysis, a preprocessing procedure was performed. Firstly, the genes obtained from three data were intersected to gain the common genes. Then, KEGG pathways containing <5 genes or >100 genes were removed, because pathways with too many genes might be too complex to understand, and pathways with too few genes may not have sufficient biological content. Finally, a total of 6919 genes, 888 drugs, and 281 pathways were selected for subsequent analysis.
iPAS For RA
In this section, the pathway levels in each sample were calculated using iPAS algorithm by making use of the accumulated normal data. In this study, 15 normal control samples were combined and regarded as references (nRef). For individual RA cases, an uniformly normalization was performed after combining the single RA data with all nRef samples.
The gene expression value of individual RA sample was standardized by mean and standard deviation (SD) of the reference. For each gene a, we calculated the gene expression level as follows:
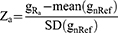
where gRa represented the expression value of gene a in an individual RA subject, mean(gnRef) stood for the mean expression value of genes a in all nRef cases, SD(gnRef) stood for SD of the reference.
To evaluate the iPAS by nRef, Average Z algorithm was employed, which presented well in highlighting pathway aberrance and in revealing clinical importance.21 A vector Z = (z1, z2, …, zn) represented the expression status of a pathway, where za stood for the standardized expression value of the a-th gene, and n represented the gene number in the specific pathway. The iPAS value of a pathway was calculated as follows:
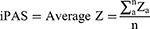
Then, the expression matrix (281 pathways × 18 RA subjects) was obtained for each pathway in each individualized RA subject from the nRef.
The pathway statistics for each individual RA subject was calculated by wilcoxon-test and false discovery rate (FDR) was used to adjust the p-value. The pathways with p-value <0.05 were defined as differential pathways.
iPAS For Drugs
The CMap contains more than 7000 gene expression profiles for 1309 compounds. Each drug presents a specific drug-induced gene expression changes of human cells, enabling us to identify the pathway aberrance. After data preprocessing, a total of 6919 genes, 888 drugs, and 281 pathways were selected for subsequent analysis. To identify the drug-induced pathway aberrance, iPAS algorithm was utilized to estimate the pathway levels. For each drug, we calculated the specific drug-induced iPAS status of each pathway using Average Z algorithm. Then, the expression matrix (281 pathways × 888 drugs) was obtained for each pathway in each drug.
Prediction Of Candidate Drugs
After the above treatment, we obtained RA-induced pathway aberrances and drug-induced pathway aberrances, respectively. Then, we systematically estimate the similarities between RA-induced pathway aberrances and drug-induced pathway aberrances using a to select drugs that might mimic or suppress RA. Prior to similarity analysis, we firstly built Prototype Ranked Lists (PRLs)25 by merging all the samples corresponding to the same drug, after converting iPAS values to ranks (the iPAS value was used as a ranking procedure in our analysis). The expression matrix (281 pathways × 888 drugs) of PRLs was obtained for further analysis.
Next, the pathway-drug PRL matrix was converted to a subject-oriented matrix. Here, a rank-based pattern-matching Enrichment Score (ES) strategy that was based on the weighted Kolmogorov–Smirnov (KS) statistic in Drug Set Enrichment Analysis (DSEA)26 was employed to perform the converted procedure. Given a PRL x and an RA subject y, the ESxy was calculated through DSEA approach. The KS-weighted ES could quantitatively measure the enrichment of signatures in the top/bottom ranked region. The ES value gives a range from 0 to 1. ES value tending to 1 indicates complete similarity and the value tending to 0 indicates the complete opposite. Finally, we built an ES matrix (888 drugs × 18 RA samples), the row corresponded to drug and the column represented RA subjects.
By calculating the ES values, the drugs tending to mimic or suppress RA were quantified. Based on the drug-subject matrix, we sorted each row x according to the ESxy values of the drug x across the y = 1, …, Y RA subjects, and obtained a rank-based drug matrix R. Given an element Rxy in R, it represented the rank of drug x according to its effect on RA subject y. In this case, the ES could sign whether a drug was a mimic or inhibitor in the development of RA.
The significance of a drug for RA subjects was assessed by applying a nonparametric, rank-based procedure. For each disease subject y, the rank value of drug x was defined as:
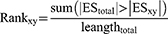
The larger the rank value, the greater the likelihood of a suppressant; the smaller the rank value, the greater the likelihood of a good mimic. In the present study, the top 1% drugs with larger rank value were predicted as therapeutic drugs, and the top 1% drugs with smaller rank value could be considered as good mimics.
Results
iPAS For RA
In the present study, 15 healthy subjects were denoted as nRefs (reference) of 18 subjects diagnosed RA. The genes were subjected to quantile normalization to evaluate the gene-level statistics. Meanwhile, a total of 281 pathways were screened from the KEGG pathway database after data preprocessing. By iPAS algorithm, we obtained the pathway aberrance scores of individual RA subjects. Using the mean value of iPAS as the pathway aberrance level of RA, 170 pathways were up-regulated and 111 pathways were down-regulated in RA. The conditions of the altered pathways in all RA subjects were elucidated by individual pathway analysis. The pathway statistics for each individual was tested by wilcoxon-test and the p-value was adjusted by FDR. Under p-value <0.05, a total of 4 pathways were regarded as differential pathways (Table 1). In our study, cardiac muscle contraction pathway showed the highest altered frequency, which altered in 14 of 18 RA subjects, followed by amoebiasis, amino sugar and nucleotide sugar metabolism, and protein processing in endoplasmic reticulum pathway.
 |
Table 1 The Differential Pathways In Rheumatoid Arthritis Based On The Individualized Pathway Aberrance Score |
iPAS For Drugs
Similar to the identification of pathway aberrance in RA, we analyzed the function aberrance induced by each drug in CMap using iPAS algorithm. For each drug, we calculated the specific gene expression profiles to detect the drug-induced iPAS status of each pathway, using untreated human cells as nRefs (reference). After data preprocessing, we obtained 888 drugs from CMap database and 281 pathways from KEGG database for further analysis. After drug iPAS analysis, we obtained a 281 pathways × 888 drugs matrix.
Prediction Of Candidate Drugs For RA
Based on the iPAS matrix of RA and drugs, a KS-weighted ES analysis was implemented to compare similarities between RA-induced pathway aberrances and drug-induced pathway aberrances. Then, a non-parametric, rank-based procedure was employed to yield a rank list to select the drug candidates that might mimic or treat RA. The pathway profiles for 888 drugs were merged into a specific PRLs by ranking the aberrant pathways. Then, pathway-oriented drug matrix was converted into an RA subject-oriented drug matrix by measuring the ES values. After that, each drug was given a specific ES value for each RA subject. A drug with high ES values indicated that the drug showed related genomic response to RA. The candidate drugs for RA subjects were assessed by applying the rank-based procedure, and the drug-subject matrix was sorted row-wise to select RA-related drugs from the most inhibiting one to the most mimicking one. Here, we identified 9 candidate drugs for RA treatment, as shown in Table 2. Also, nine good mimics were identified based on the similarity of pathway profiles to disease (Table 3). Some candidates with current indications to treat other diseases might be repurposed to treat RA.
 |
Table 2 The Candidate Therapeutic Drugs Of Rheumatoid Arthritis |
 |
Table 3 The Good Mimics Of Rheumatoid Arthritis |
Discussion
In response to the high attrition rates in the traditional drug development process, drug repositioning which recaptures marketed drugs for new indications has attracted the attention from pharmaceutical companies and medical researchers. Ashburn and Thor indicated that drug repositioning might shorten the time of drug development from 10–17 years to 3–12 years.27 Previous studies have proposed numerous methods to build predictive models and some have shown promising results.10,12,28,29 These emerging technologies enable investigators to identify candidate drugs that will prevent this disease and its complications. For example, Zhang et al30 presented a drug repositioning strategy based on “omics” data mining to screen candidates for new indications in diabetes treatment, and successfully identified 9 drugs that might have the potential to treat diabetes.
Existing drug repositioning methods mainly focus on discovering candidate drugs for a kind of disease, and are not suitable for predicting candidate drugs for an individual sample. The drug response heterogeneity makes the raise of new strategies that target genotypically well-characterized subpopulations of patients, instead of targeting huge populations. This conversion drives the researchers focus to settle personalized pathway profile by which the disease works and how to intervene on it. In this study, we proposed a computational method to predict candidate drugs from CMap database for RA, in a personalized way, contributing to revealing the molecular mechanisms and the future application of custom therapeutic decisions.
In the present study, we first identify the pathway aberrance of individual RA subjects using iPAS algorithm, and a total of 4 differential pathways were identified. It is well known that RA is a kind of autoimmune disorder. Our study identified 4 differential pathways (cardiac muscle contraction, amoebiasis, amino sugar and nucleotide sugar metabolism, and protein processing in endoplasmic reticulum) in RA. Among them, cardiac muscle contraction pathway showed the highest altered frequency, which altered in 14 of 18 RA subjects. Previous studies revealed that RA patients had an increased risk of premature death compared with the general population, mainly due to cardiovascular disease, and the two diseases shared genetic and environmental risk factors.31,32 Our result that identified cardiac muscle contraction as a differential pathway might affirm this declaration.
Although, a rapid evolution of the care and treatment of patients with RA has contributed to a decrease in disease and disease-related complications. Efficient and safe drugs are still pressing problem. By drug repositioning, several small molecules that might suppress RA were identified, which might be critically important for disease prevention and early treatment. Among these candidate drugs, celecoxib had a current drug indication of RA, which is a COX-2 selective nonsteroidal anti-inflammatory drug used to treat various forms of arthritis.33 The candidate parthenolide showed a current drug indication of anti-inflammatory. López-Franco et al34 indicated that parthenolide could modulate the NF-κB-mediated inflammatory responses during vascular damage. Moreover, parthenolide has a variety of reported in vitro biological activities, such as blocking lipopolysaccharide-induced osteolysis,35 inducing apoptosis of leukemia cells,36 and activity against a parasite Leishmania amazonensis.37 The candidate drug harmine was reported to promote differentiation of osteoblasts38 and chondrocytes,39 and inhibit osteoclastogenesis.40 Suloctidil and prenylamine both showed a current drug indication of vasodilator. Unfortunately, they were withdrawn from market due to liver toxicity and cardiac arrhythmias, respectively. The other candidate drugs predicted by our method had current indications to treat other diseases and might complement the drug group for RA treatment.
Given these screened potential drug for RA treatment, previous literature have reported that some of these drugs could exert certain relieved effect on RA. For instance, Danazol has been identified that treatment of it attenuates refractory autoimmune thrombocytopenia in rheumatic diseases successfully.41,42 Some studies have demonstrated that piperlongumine relieves RA through expansion of myeloid-derived suppressor cells (MDSCs) and the inhibition of the Th17 response and activation of fibroblast-like synoviocytes (FLS)43 represses dendritic cell maturation by decreasing production of reactive oxygen species,44 and suppresses proliferation, migration and invasion of FLS45 via animal model and patient samples. Celecoxib has been approved to treat RA and osteoarthritis (OA);46,47 it, combined with Cilostazol, impedes proinflammatory factors in FLS of RA patients.48 These results may provide more favorable evidence for the clinical use of these drugs.
Meanwhile, we predicted nine mimics based on the similarity of pathway aberrances between disease and drugs. In this study, the drugs with higher ES values showed more similar to disease in the matter of pathway aberrances. Theoretically, it happened that the drugs with high ES values might mimic the disease at the molecular level. While it may not make sense to consider that the pathway profiles of one drug is positively related to those of one disease from the biological view. Thus, this study focused on the candidate drugs for RA treatment in a personalized way.
Conclusions
In the present study, we predicted several candidate drugs for RA treatment, and conducted drug discovery in a personalized way contributing to the future application of custom therapeutic decisions.
Acknowledgments
This study was funded by the grants from Science and Technology Foundation, Health and Family Planning Commission of Guizhou Province, China (No. gzwjkj2015-1-025), and Joint Project of Guizhou Provincial Department of Science and Technology and Guizhou Provincial People’s Hospital, China (LH7006). Ru-Yin Hu and Xiao-Bin Tian are equal contributors and co-first authors for this study.
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Firestein GS. Evolving concepts of rheumatoid arthritis. Nature. 2003;423(6937):356–361. doi:10.1038/nature01661
2. Alamanos Y, Drosos AA. Epidemiology of adult rheumatoid arthritis. Autoimmun Rev. 2005;4(3):130–136. doi:10.1016/j.autrev.2004.09.002
3. Smolen JS, Breedveld FC, Burmester GR, et al. Treating rheumatoid arthritis to target: 2014 update of the recommendations of an international task force. Ann Rheum Dis. 2016;75(1):3–15. doi:10.1136/annrheumdis-2015-207524
4. Singh JA, Saag KG, Bridges SL
5. Smolen JS, Landewe R, Breedveld FC, et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2013 update. Ann Rheum Dis. 2014;73(3):492–509. doi:10.1136/annrheumdis-2013-204573
6. Hurle MR, Yang L, Xie Q, Rajpal DK, Sanseau P, Agarwal P. Computational drug repositioning: from data to therapeutics. Clin Pharmacol Ther. 2013;93(4):335–341. doi:10.1038/clpt.2013.1
7. Boguski MS, Mandl KD, Sukhatme VP. Drug discovery. Repurposing with a difference. Science. 2009;324(5933):1394–1395. doi:10.1126/science.1169920
8. Cheng F, Liu C, Jiang J, et al. Prediction of drug-target interactions and drug repositioning via network-based inference. PLoS Comput Biol. 2012;8(5):e1002503. doi:10.1371/journal.pcbi.1002503
9. Luo H, Chen J, Shi L, et al. DRAR-CPI: a server for identifying drug repositioning potential and adverse drug reactions via the chemical-protein interactome. Nucleic Acids Res. 2011;39(Web Server issue):W492–W498. doi:10.1093/nar/gkr299
10. Yang L, Agarwal P. Systematic drug repositioning based on clinical side-effects. PLoS One. 2011;6(12):e28025. doi:10.1371/journal.pone.0028025
11. Lamb J, Crawford ED, Peck D, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. doi:10.1126/science.1132939
12. Li J, Zhu X, Chen JY. Building disease-specific drug-protein connectivity maps from molecular interaction networks and PubMed abstracts. PLoS Comput Biol. 2009;5(7):e1000450. doi:10.1371/journal.pcbi.1000450
13. Dopazo J. Genomics and transcriptomics in drug discovery. Drug Discov Today. 2014;19(2):126–132. doi:10.1016/j.drudis.2013.06.003
14. Okada Y, Wu D, Trynka G, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506(7488):376–381. doi:10.1038/nature12873
15. Kurko J, Besenyei T, Laki J, Glant TT, Mikecz K, Szekanecz Z. Genetics of rheumatoid arthritis – a comprehensive review. Clin Rev Allergy Immunol. 2013;45(2):170–179. doi:10.1007/s12016-012-8346-7
16. Mathews RJ, Robinson JI, Battellino M, et al. Evidence of NLRP3-inflammasome activation in rheumatoid arthritis (RA); genetic variants within the NLRP3-inflammasome complex in relation to susceptibility to RA and response to anti-TNF treatment. Ann Rheum Dis. 2014;73(6):1202–1210. doi:10.1136/annrheumdis-2013-203276
17. Teixeira VH, Olaso R, Martin-Magniette ML, et al. Transcriptome analysis describing new immunity and defense genes in peripheral blood mononuclear cells of rheumatoid arthritis patients. PLoS One. 2009;4(8):e6803. doi:10.1371/journal.pone.0006803
18. Walsh AM, Whitaker JW, Huang CC, et al. Integrative genomic deconvolution of rheumatoid arthritis GWAS loci into gene and cell type associations. Genome Biol. 2016;17:79. doi:10.1186/s13059-016-0948-6
19. Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402(6761 Suppl):C47–C52. doi:10.1038/35011540
20. Glazko GV, Emmert-Streib F. Unite and conquer: univariate and multivariate approaches for finding differentially expressed gene sets. Bioinformatics. 2009;25(18):2348–2354. doi:10.1093/bioinformatics/btp406
21. Ahn T, Lee E, Huh N, Park T. Personalized identification of altered pathways in cancer using accumulated normal tissue data. Bioinformatics. 2014;30(17):i422–i429. doi:10.1093/bioinformatics/btu449
22. Gautier L, Cope L, Bolstad BM, Irizarry RA. AFSFY – analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi:10.1093/bioinformatics/btg405
23. Dai M, Wang P, Boyd AD, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33(20):e175. doi:10.1093/nar/gni179
24. Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–D462. doi:10.1093/nar/gkv1070
25. Allan J, Leuski A, Swan R, Byrd D. Evaluating combinations of ranked lists and visualizations of inter-document similarity. Information Process Manage. 2001;37:435–458. doi:10.1016/S0306-4573(00)00056-X
26. Napolitano F, Sirci F, Carrella D, di Bernardo D. Drug-set enrichment analysis: a novel tool to investigate drug mode of action. Bioinformatics. 2016;32(2):235–241. doi:10.1093/bioinformatics/btv536
27. Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3(8):673–683. doi:10.1038/nrd1468
28. Kotelnikova E, Yuryev A, Mazo I, Daraselia N. Computational approaches for drug repositioning and combination therapy design. J Bioinform Comput Biol. 2010;8(3):593–606. doi:10.1142/S0219720010004732
29. Keiser MJ, Setola V, Irwin JJ, et al. Predicting new molecular targets for known drugs. Nature. 2009;462(7270):175–181. doi:10.1038/nature08506
30. Zhang M, Luo H, Xi Z, Rogaeva E. Drug repositioning for diabetes based on ‘omics’ data mining. PLoS One. 2015;10(5):e0126082. doi:10.1371/journal.pone.0126082
31. Nurmohamed MT, Heslinga M, Kitas GD. Cardiovascular comorbidity in rheumatic diseases. Nat Rev Rheumatol. 2015;11(12):693–704. doi:10.1038/nrrheum.2015.112
32. Skeoch S, Bruce IN. Atherosclerosis in rheumatoid arthritis: is it all about inflammation? Nat Rev Rheumatol. 2015;11(7):390–400.
33. McCormack PL. Celecoxib: a review of its use for symptomatic relief in the treatment of osteoarthritis, rheumatoid arthritis and ankylosing spondylitis. Drugs. 2011;71(18):2457–2489. doi:10.2165/11208240-000000000-00000
34. Lopez-Franco O, Hernandez-Vargas P, Ortiz-Munoz G, et al. Parthenolide modulates the NF-kappaB-mediated inflammatory responses in experimental atherosclerosis. Arterioscler Thromb Vasc Biol. 2006;26(8):1864–1870. doi:10.1161/01.ATV.0000229659.94020.53
35. Yip KH, Zheng MH, Feng HT, Steer JH, Joyce DA, Xu J. Sesquiterpene lactone parthenolide blocks lipopolysaccharide-induced osteolysis through the suppression of NF-kappaB activity. J Bone Miner Res. 2004;19(11):1905–1916. doi:10.1359/JBMR.040919
36. Zunino SJ, Ducore JM, Storms DH. Parthenolide induces significant apoptosis and production of reactive oxygen species in high-risk pre-B leukemia cells. Cancer Lett. 2007;254(1):119–127. doi:10.1016/j.canlet.2007.03.002
37. Tiuman TS, Ueda-Nakamura T, Garcia Cortez DA, et al. Antileishmanial activity of parthenolide, a sesquiterpene lactone isolated from Tanacetum parthenium. Antimicrob Agents Chemother. 2005;49(1):176–182. doi:10.1128/AAC.49.11.176-182.2005
38. Yonezawa T, Lee JW, Hibino A, et al. Harmine promotes osteoblast differentiation through bone morphogenetic protein signaling. Biochem Biophys Res Commun. 2011;409(2):260–265. doi:10.1016/j.bbrc.2011.05.001
39. Hara ES, Ono M, Kubota S, et al. Novel chondrogenic and chondroprotective effects of the natural compound harmine. Biochimie. 2013;95(2):374–381. doi:10.1016/j.biochi.2012.10.016
40. Egusa H, Doi M, Saeki M, et al. The small molecule harmine regulates NFATc1 and Id2 expression in osteoclast progenitor cells. Bone. 2011;49(2):264–274. doi:10.1016/j.bone.2011.04.003
41. Dasgupta B, Grahame R. Treatment with danazol for refractory thrombocytopenia in rheumatoid arthritis. Br J Rheumatol. 1989;28(6):550–552. doi:10.1093/rheumatology/28.6.550
42. Blanco R, Martinez-Taboada VM, Rodriguez-Valverde V, Sanchez-Andrade A, Gonzalez-Gay MA. Successful therapy with danazol in refractory autoimmune thrombocytopenia associated with rheumatic diseases. Br J Rheumatol. 1997;36(10):1095–1099. doi:10.1093/rheumatology/36.10.1095
43. Sun J, Xu P, Du X, Zhang Q, Zhu Y. Piperlongumine attenuates collagen-induced arthritis via expansion of myeloid-derived suppressor cells and inhibition of the activation of fibroblast-like synoviocytes. Mol Med Rep. 2015;11(4):2689–2694. doi:10.3892/mmr.2014.3001
44. Xiao Y, Shi M, Qiu Q, et al. Piperlongumine suppresses dendritic cell maturation by reducing production of reactive oxygen species and has therapeutic potential for rheumatoid arthritis. J Immunol. 2016;196(12):4925–4934. doi:10.4049/jimmunol.1501281
45. Xu S, Xiao Y, Zeng S, et al. Piperlongumine inhibits the proliferation, migration and invasion of fibroblast-like synoviocytes from patients with rheumatoid arthritis. Inflammation Res. 2018;67(3):233–243. doi:10.1007/s00011-017-1112-9
46. Fidahic M, Jelicic Kadic A, Radic M, Puljak L. Celecoxib for rheumatoid arthritis. Cochrane Database Syst Rev. 2017;6:Cd012095.
47. Krasselt M, Baerwald C. Celecoxib for the treatment of musculoskeletal arthritis. Expert Opin Pharmacother. 2019;20(14):1689–1702. doi:10.1080/14656566.2019.1645123
48. Lee YS, Lee SY, Park SY, Lee SW, Hong KW, Kim CD. Cilostazol add-on therapy for celecoxib synergistically inhibits proinflammatory cytokines by activating IL-10 and SOCS3 in the synovial fibroblasts of patients with rheumatoid arthritis. Inflammopharmacology. 2019. doi:10.1007/s10787-019-00605-5
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
