Back to Journals » The Application of Clinical Genetics » Volume 11
Independent of DAZL-T54A variant and AZF microdeletion in a sample of Egyptian patients with idiopathic non-obstructed azoospermia
Authors El Shafae MM, Sabry JH , Behiry EG , Sabry HH, Salim MA, Fayez AG
Received 28 November 2017
Accepted for publication 12 April 2018
Published 19 July 2018 Volume 2018:11 Pages 81—87
DOI https://doi.org/10.2147/TACG.S158297
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Martin Maurer
Mohammed M El Shafae,1 Jehan H Sabry,1 Eman G Behiry,1 Hanan H Sabry,2 Mona A Salim,1 Alaaeldin G Fayez3
1Department of Clinical and Chemical Pathology, Benha Faculty of Medicine, Benha University, Benha, Egypt; 2Department of Dermatology, Venereology and Andrology, Benha Faculty of Medicine, Benha University, Benha, Egypt; 3Department of Molecular Genetics and Enzymology, Human Genetics and Genome Research Division, National Research Centre, Cairo, Egypt
Background: The microdeletion events that occur in the Y chromosome-azoospermia factor (AZF) region may lead to dyszoospermia. Also, the deleted azoospermia (DAZ) gene on AZFc and autosomal deleted azoospermia like gene (DAZL) are suggested to represent impairment, so it is interesting to determine the independency pattern of the AZF region and DAZL gene in azoospermic patients.
Aim: To study the molecular characterization of AZFc and DAZL in 64 idiopathic non-obstructed azoospermia patients and 30 sexually reproductive men.
Methods: SYBR Green I (Q-PCR) and AZF-STS analysis was used for DAZ gene, and SNV-PCR and confirmative Sanger sequencing for DAZL gene.
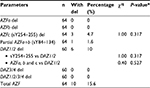
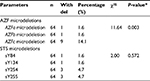
Results: The present study observed that 15.6% had AZFc microdeletion, out of which 10% had DAZ1/2 deletion, and no T54A variant in the DAZL gene was found.
Conclusion: In the current work, the novelty is that spermatogenic impairment phenotype, present with AZFc microdeletions, is independent of the T54A variant in the DAZL gene, and AZFc microdeletions could be a causative agent in spermatogenic impairment.
Keywords: male infertility, azoospermia, AZF, DAZL, deletion
Introduction
Worldwide, infertility is estimated to affect about 186 million people (from 8 to 12% of reproductive-aged couples), and more than half of all cases of global childlessness are due to male infertility.1,2 It is estimated that, in about 30% of cases, male infertility is due to a genetic disorder such as aneuploidy, structural chromosomal abnormalities, DNA damage, and gene mutations, including a variety of newly discovered genes.3
There has been an intensive search for genetic causes of male infertility, of which spermatogenic failure is the most common form. Screening with markers on the long arm of the human Y chromosome has detected Yq microdeletions in 5–15% of males with spermatogenic failure. Among cases with Yq microdeletions, deletion involving the azoospermia factor (AZF) region in the Y chromosome has been discovered to be a frequent genetic cause associated with male infertility.4 The AZFc region is particularly interesting, as approximately 80% of AZF microdeletions occur in this region, and most of them result in entire DAZ (deleted in azoospermia) gene deletion.5 The DAZ gene has four copies and most commonly encodes an RNA-binding protein exclusively in testicular tissue.6,7 Studies have demonstrated that the DAZ gene plays an important role in spermatogenesis.8,9 Although most deletions involve a deletion of all four DAZ genes, an absence of only two of the DAZ genes is also associated with impaired spermatogenesis.10 Detailed analysis of the AZFc region using new molecular non-repeating sequence-tagged site (STS) markers has confirmed the existence of three such microdeletions, namely, gr/gr, b1/b3, and b2/b3.11,12 The most prevalent partial deletion, the gr/gr deletion, is caused by recombination between amplicons g and r (g1, r1, and/or r2, with their respective homologous amplicons g2, r3, and/or r4).13,14
The DAZ gene has an autosomal homolog, DAZL (DAZ-like), on chromosome 3p24. It is highly homologous to the DAZ gene, with 83% similarity in the coding region of the cDNA. Both genes encode RNA-binding proteins. It is believed to play a role in spermatogenesis.15–17
DAZL gene encoding RNA binding protein is specifically expressed in germ cells of male and female and targets Tpx-1.18 Tex19.1, Sycp3, and DDX4 transcripts in its 3′UTR region.19 In Taiwanese men, but not in Caucasians, who ranged from hypospermatogenesis and maturation arrest to Sertoli cell-only syndrome, Teng et al20 identified a c.386G>A transition in exon 3 of the DAZL gene which led to p.T54A substitution in the RNA recognition motif domain in 7.39% of patients. p.T54A was not detected in populations from Germany, Italy, Japan, Northern China, or Western India. Because that p.T54A variant of the DAZL gene has not been tested on infertile Egyptian men, and DAZ copy number variations is considered a main key in spermatogenesis, it is reasonable to investigate conjoining DAZ and DAZL genotyping in male infertility.
We aimed to study microdeletions in the AZF region and copy number variations in the DAZ region and also study p.T54A variant of DAZL gene in idiopathic non-obstructed azoospermic (NOA) Egyptian patients.
Subjects and methods
This study was approved by the Medical Ethical Committee of Benha Faculty of Medicine, Benha University, and the Medical Ethical Committee of the National Research Centre (Egypt) according to the “World Medical Association Declaration of Helsinki.” Written informed consent was signed by all participants.
This case-controlled study was conducted on 64 NOA Egyptian patients (with age ranging from 20–47) years who were examined in the Department of Dermatology and Andrology, Benha Faculty of Medicine, Benha University, and National Research Centre, between January and December 2016.
Patients with spermatogenic impairment due to causes, such as obstruction of the vas deferens, history of and/or active orchitis, hyperprolactinemia, hypogonadotropic hypogonadism, previous chemo- or radiotherapy, or a history of unilateral and bilateral cryptorchidism and varicocele were excluded. The patients were evaluated for karyotype abnormalities, and those showing chromosomal abnormalities were excluded.
All patients underwent comprehensive surveillance, including a detailed history taking, physical examination, at least two semen analyses, endocrinology profiles testing (LH, FSH, prolactin [PRL], and testosterone). Semen samples were collected by masturbation after 3–5 days of abstinence. The diagnosis of azoospermia was established by pellet analysis after semen centrifugation that was repeated at least twice to confirm azoospermia. In patients with highly suspected non-obstructive azoospermia, bilateral testicular fine needle aspiration cytological analysis were done. Non-obstructive azoospermia was defined as: 1) spermatogenic defects in the testicular cytology (such as hypospermatogenesis, maturation arrest, and Sertoli cell-only syndrome) or 2) elevated serum FSH level, total testicular volume less than 30 ml. Semen analysis was performed according to the standard methods outlined by the World Health Organization.21
Thirty age-matched proven fertile men with a normal semen analysis and karyotype were recruited as controls. The control subjects were husbands of women who received regular prenatal care at the University hospital. All control individuals had fathered at least one child during the last 3 years and never had any sexual abnormality.
Molecular investigations
I-a-AZF-STS analysis
Blood samples were collected using Na2EDTA as an anticoagulant inside vacutainer sterile tubes. DNA was isolated from peripheral blood leukocytes followed by AZF-STS analysis using six AZFc-specific STSs (sY84, sY86, sY127, sY134, sY245, and sY255) according to the European Academy of Andrology and the European Molecular Genetics Quality Network (EAA/EMQN) guidelines.22 This guideline indicated that the use of these six STS loci are most relevant to the reported Y-chromosome microdeletions cases. An STS was considered absent only after at least two amplification failures in the presence of successful amplification of control (SRY-sY14). AZF-STS microdeletion screening was done by two multiplex polymerase chain reactions (PCRs), each covering the three AZF regions.
I-b-Copy number estimation of DAZ genes using SYBR Green I (Q-PCR)
We estimated copy numbers of DAZ genes using KAPA SYBR FAST qPCR Master mix (2X) (Code; KM4100, Kapa Biosystem, Boston, MA, USA), the primer sequences are shown in Table 1. Four copied STS sY587 located in intron 10 of the DAZ gene were chosen for quantification. All runs were carried out in duplicate, with the calibrator sample containing the four DAZ genes and a reference sample. qPCR assay was performed using 40 cycles at denaturation 95°C for 8 s, annealing at 58°C for 20 s, extension at 72°C for 3 s, followed by a dissociation step from 40°C to 85°C according to the Roch Light Cycler 480 II instrument guideline. The data were analyzed using the comparative Ct (∆∆Ct) relative quantitation assay method.23
  | Table 1 DAZ gene fragments after DraI digestion Abbreviations: DAZ, deleted azoospermia; STS, sequence-tagged site; bps, base pairs. |
I-c-Characterization of partial DAZ deletion using SNV-PCR
To determine which type of DAZ gene is deleted, we carried out single-nucleotide variants (SNVs) PCR analysis using sY587/DraI PCR-restriction fragment length polymorphism (RFLP). The digested products (Table 2) were run on a 3% agarose gel containing ethidium bromide and visualized by BioRad Gel doc instrument.
  | Table 2 The designed primers sequence for exon 3 of the DAZL gene and AZF spanning primers Abbreviations: DAZL, deleted azoospermia like gene; AZF, azoospermia factor. |
II-a-Genotyping of T54A variant in DAZL gene by PCR-RFLP
Genotyping of T54A variant in DAZL gene was performed in 25 μL final volume reaction mix, containing up to 1 µg of genomic DNA. We designed specific PCR primers (Table 1) covering exon 3 (based on GRCh37.p13 Primary Assembly) using NCBI Primer-BLAST tool.24 The amplified product was 262 bps in length. Amplicons were digested using the restriction enzyme AluI (New England Biolabs, Ipswich, MA, USA). The restriction fragments were run on a 4% agarose gel. The normal allele is cut into three restriction fragments of 142, 115, and 5 bps, whereas the polymorphism A→G creates an AluI restriction site (AGCT) giving four fragments of 129, 115, 13, and 5 bps.
II-b-Confirmative Sanger sequencing for exon 3 of DAZL gene
To confirm the restriction enzyme AluI results, amplicon of exon 3 was followed by direct Sanger Sequencing using the BigDye Terminator Cycle Sequencing kit (Perkin-Elmer Inc., Waltham, MA, USA) on the ABI3730XL sequencer in Macrogen Inc. (South Korea) (http://dna.macrogen.com).
Statistical methods
The obtained data were tabulated and analyzed using SPSS version 16 software (SPSS Inc., Chicago, IL, USA). Categorical data were presented as numbers and percentages. A P-value <0.05 was considered statistically significant and <0.01 was considered highly statistically significant. Chi square test (χ2), odds ratios (ORs), and the corresponding 95% CI were calculated when applicable.
Results
The mean age of subjects was 31.4±6.1 years (range=20–47 years). Both studied groups showed normal hormonal mean levels (follicle stimulating hormone [FSH], luteinizing hormone [LH], and testosterone) with a non-significant statistical difference. qPCR showed that ∆∆Ct normal references for the four DAZ copies (b2/b4) is 4 (3.6–4.8), and for the two copies (gr/gr, b1/b3, and g1/g3) is 2 (1.6–2.2). DAZ-copy number variant (CNV) by SNV-PCR was done in cases that showed reducing DAZ gene numbers by real-time PCR only. PCR-RFLP showed that, among 60 azoospermic cases, six cases had DAZ1/2 deletion (6/60, 10%) (Figure 1). None of the exon 3 PCR products of 64 patients and 30 controls showed a mutant digestion pattern in DAZL (Figure 2). The obtained DAZL digestion patterns were confirmed by bidirectional Sanger sequencing, which showed no clear A→G transition (Figure 3). All normospermic fertile men (control group) had no detected AZF deletions using the same technique. The full results are shown in Tables 3 and 4.
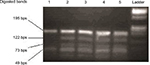  | Figure 1 Agarose gel electrophoresis for restriction enzyme assay by DraI showed that case no 1 had an absent 122 bps fragment, indicating that DAZ1/2 were deleted. Abbreviation: bps, base pairs. |
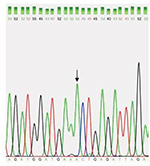  | Figure 3 Partial DNA sequence chromatogram for exon 3 of the DAZL gene shows normal A allele (arrow). Abbreviation: DAZL, deleted azoospermia like gene. |
Discussion and conclusion
Infertility is a disease that affects about one in six couples at reproductive age. Approximately half of the infertile cases are due to male factors, including abnormal semen parameters and azoospermia.21,25
The frequency and type of Y chromosome microdeletions varied according to ethnic, regional differences, selection criteria for patient samples, or methodological differences.26 Reports on the presence of Y chromosome microdeletions in infertile men range from 0.23% to 34.5%, regardless of the number of STSs used, this wide range may be attributed to population/ethnic variances, selection criteria of the patients, methodological aspects, and environmental factors.27–29 Our results reported AZF-Y chromosome microdeletion in 15.6%, where 4.7% presented in sY245 and 255 together, 1.6% presented in sY84 and 134 together, and 10% (6/60) presented in DAZ1/2 deletion.
To our knowledge, there are few studies which determined copy number variation of DAZ genes by quantitative method such as real-time PCR technique. Also, T54A variant in multiple populations has long been investigated. However, no studies have specifically examined this variant in infertile men in an Egyptian population. Our study investigated the DAZ copy number variations using relative quantitative real-time PCR followed by SNV-PCR to analyze the variations of DAZ genes in those patients and investigated the T54A variant in exon 3 of DAZL gene using RFLP-PCR.
Shimizu et al30 proposed that approximately 10% of non-obstructive azoospermia patients are positive for Y chromosome AZF microdeletion. Liu et al31 showed that infertile non-obstructive azoospermia and severe oligospermia patients had an increased tendency to AZFc partial deletions (7.40%) and AZFc deletions (4.14%). Others studies showed that, in the Caucasian population, 15% of idiopathic azoospermia cases had deletion of either four DAZ genes or DAZ1/DAZ2 in the AZFc region, and 8.8% of Chinese azoospermic patients had complete deletion of DAZ genes, and DAZ1/DAZ2 deletion was confirmed in 8.3% of azoospermic patients.32–34 Our results showed a frequency of 10% of studied NOA had DAZ1/2 deletion and 4.7% had complete AZFc deletion, whereas other studies could not confirm the relationship between AZF microdeletions and male infertility.35,36
A number of previous studies pointed that the gr/gr deletion and the g1/g2, which remove DAZ copies 1 and 2, represented a risk factor for spermatogenic damage. In contrast, the b2/b3, gr/gr, and g1/g3 deletions, which remove DAZ copies 3 and 4, seemed to have no or little effect on fertility.37 In our results, no DAZ3/4 deletion was detected.
According to our knowledge, the Egyptian population was not tested for p.T54A variant, so the allele frequency for this variant in those populations is still unknown, and this is the first report about the T54A variant in a non-Taiwan Chinese population.
Stratified analysis was performed by Zhang et al38 via meta-analysis of 13 case–control studies, including 2556 cases and 1997 controls, showed a strong association between p.T54A polymorphism and male infertility in Asians.
Tüttelmann et al39 reported, through a meta analysis study, that the p.T54A mutation was never found in non-Chinese populations and seems to be a factor associated with male infertility only in Taiwan.
Regarding the fact that the p.T54A variant of DAZL gene has not been studied in Egyptian infertile men, yet, we screened A and G alleles for A360G variant of the DAZL gene using PCR-RFLP in 170 alleles of Egyptian infertile patients and 100 alleles of controls in order to study the allelic frequency of this variant in a sample of the Egyptian population. We did not find G allele in the patients or in the controls. Additional Direct Sanger sequencing for three samples with forward primer also showed A allele only.
Bartoloni et al40 described that sequencing for A>G transition variant with the reverse primer was better from the forward one, because the forward primer led to a background which looked like the A>G transition that was absent in sequences from the reverse primer. However, our results showed that sequencing A>G transition variant using the forward primer was clear, without a noisy background (Figure 2).
Chen et al41 reported that partial AZFc deletions are independent of the variations in DAZL. In our results, no coexistence pattern was found between the T54A variant of DAZL gene and AZF defect in spermatogenic impairment in men with azoospermia.
Y chromosome studies could serve as a predictive factor in probability of sperm retrieval. So, the Y chromosome studies are important. Also, AZF microdeletion may be passed onto the next male generation; hence, it is important to screen specific DNA sequences on the Y chromosome before ICSI. Liu et al31 pointed out that detection of Y chromosome microdeletions is of great use for guiding clinical diagnosis, selecting treatment schemes, and reducing the incidence of this genetic disease .
We can conclude that 1) our study provides further evidence that partial deletions of the AZFc region, especially DAZ1/2 deletion, are a risk factor for spermatogenesis impairment rather than DAZ3/4 deletion, and this agrees with what was reported by Fayez et al;42 2) the spermatogenic impairment phenotype of AZF microdeletions is independent of the T54A variant in the DAZL gene; and 3) the p.T54A variant may be a founder variant for the Taiwanese population or incorrect result.
Therefore, we recommend evaluating the p.T54A variant in other populations to stand on the assumption that this variant is of doubtful existence. Also, because of different potential genetic factors contributing to the spermatogenic phenotype among populations of different ethnic origins, further genetic studies are required in more populations.
The novelty of the current study is independence of the spermatogenic impairment phenotype of AZF microdeletions and T54A variant in the DAZL gene, also the absence of the T54A variant in a sample of Egyptian infertile patients.
There were several limitations to this study: 1) only patients with NOA were examined; 2) the prevalence of these genetic factors used in our study is still unknown in the Egyptian population; and 3) no familial members were included to follow the AZF microdeletion.
Acknowledgments
The authors are very grateful to patients and controls for their participation and cooperation during this study. Financial funding of this paper was provided by the authors only.
Disclosure
All authors report no conflicts of interest in this work.
References
Inhorn, MC, Patrizio P. Infertility around the globe: new thinking on gender, reproductive technologies and global movements in the 21st century. Hum Reprod Update. 2015;21(4):411–426. | ||
Ombelet W, Cooke I, Dyer S, Serour G, Devroey P. Infertility and the provision of infertility medical services in developing countries. Hum Reprod Update. 2008;14(6):605–621. | ||
Vogt PH. Molecular genetic of human male infertility: from genes to new therapeutic perspectives. Curr Pharm Des. 2004;10(5):471–500. | ||
Vogt PH, Edelmann A, Kirsch S, et al. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet. 1996;5(7):933–43. | ||
Simoni M, Bakker E, Krausz C. EAA/EMQN best practice guidelines for molecular diagnosis of Y-chromosomal microdeletions. State of the art 2004. Int J Androl. 2004;27(4):240–249. | ||
Kim B, Lee Y, Kim Y, et al. Polymorphic expression of DAZ proteins in the human testis. Hum Reprod. 2009;24(6):1507–1515. | ||
Writzl K, Zorn B, Peterlin B. Copy number of DAZ genes in infertile men. Fertil Steril. 2005;84(5):1522–1525. | ||
Fernandes AT, Fernandes S, Gonçalves R, et al. DAZ gene copies: evidence of Y chromosome evolution. Mol Hum Reprod. 2006;12(8):519–523 | ||
Reynolds N, Cooke HJ. Role of the DAZ genes in male fertility. Reprod Biomed Online. 2005;10(1):72–80. | ||
Teng YN, Chang YP, Tseng JT, et al. A single-nucleotide polymorphism of the DAZL gene promoter confers susceptibility to spermatogenic failure in the Taiwanese Han. Hum Reprod. 2012;27(9):2857–2865. | ||
Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423(6942):825–837. | ||
Repping S, Skaletsky H, Brown L, et al. Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat Genet. 2003;35(3):247–251. | ||
Kuroda-Kawaguchi T, Skaletsky H, Brown LG, et al. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet. 2001;29(3):279–286. | ||
Hucklenbroich K, Gromoll J, Heinrich M, Hohoff C, Nieschlag E, Simoni M. Partial deletions in the AZFc region of the Y chromosome occur in men with impaired as well as normal spermatogenesis. Hum Reprod. 2005;20(1):191–197. | ||
Saxena R, Brown LG, Hawkins T, et al. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nat Genet. 1996;14(3):292–299. | ||
Shan Z, Hirschmann P, Seebacher T, et al. A SPGY copy homologous to the mouse gene Dazla and the Drosophila gene boule is autosomal and expressed only in the human male gonad. Hum Mol Genet. 1996;5(12):2005–2011. | ||
Yen PH, Chai NN, Salido EC. The human autosomal gene DAZLA: testis specificity and a candidate for male infertility. Hum Mol Genet. 1996;5(12):2013–2017. | ||
Jiao X, Trifillis P, Kiledjian M. Identification of target messenger RNA substrates for the murine deleted in azoospermia-like RNA-binding protein. Biol Reprod. 2002;66(2):475–485. | ||
Reynolds N, Collier B, Maratou K, et al. Dazl binds in vivo to specific transcripts and can regulate the pre-meiotic translation of Mvh in germ cells. Hum Mol Genet. 2005;14(24):3899–3909. | ||
Teng YN, Lin YM, Sun HF, Hsu PY, Chung CL, Kuo PL. Association of DAZL haplotypes with spermatogenic failure in infertile men. Fertil Steril. 2006;86(1):129–135. | ||
World Health Organization. Examination and Processing Human Semen. Geneva: World Health Organization; 2010:1–287. | ||
Krausz C, Hoefsloot L, Simoni M, Tüttelmann F. EAA/EMQN best practice guidelines for molecular diagnosis of Y-chromosomal microdeletions: state-of-the-art 2013. Andrology. 2014;2(1):5–19. | ||
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods. 2001;25(4):402–408. | ||
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. | ||
Sen S, Pasi AR, Dada R, Shamsi MB, Modi D. Y chromosome microdeletions in infertile men: prevalence, phenotypes and screening markers for the Indian population. J Assist Reprod Genet. 2013;30(3):413–422. | ||
Krausz C, Forti G, McElreavey K. The Y chromosome and male fertility and infertility. Int J Androl. 2003;26(2):70–75. | ||
Foresta C, Moro E, Ferlin A. Y chromosome microdeletions and alterations of spermatogenesis. Endocr Rev. 2001;22(2):226–239. | ||
Mierla D, Jardan D, Stoian V. Chromosomal abnormality in men with impaired spermatogenesis. Int J Fertil Steril. 2014;8(1):35–42. | ||
Gonçalves C, Cunha M, Rocha E, et al. Y-chromosome microdeletions in nonobstructive azoospermia and severe oligozoospermia. Asian J Androl. 2017;19(3):338–345. | ||
Shimizu A, Ichikawa T, Suzuki N, et al. Microdeletions in the Y chromosome of patients with idiopathic azoospermia. Asian J Androl. 2002;4(2):111–115. | ||
Liu XG, Hu HY, Guo YH, Sun YP. Correlation between Y chromosome microdeletion and male infertility. Genet Mol Res. 2016;15(2):1–6. | ||
Yuan Y, Xiao CY, Zhang SZ, Zhang SX, Huang MK, Lin L. High risk genetic factor in Chinese patients with idiopathic male infertility; deletion of DAZ gene copy on Y chromosome. Chin Med J. 2004;117:1092–1094. | ||
Dada R, Gupta NP, Kucheria K. Yq microdeletions – azoospermia factor candidate genes and spermatogenic arrest. J Biomol Tech. 2004;15(3):176–183. | ||
Fernandes S, Huellen K, Goncalves J, et al. High frequency of DAZ1/DAZ2 gene deletions in patients with severe oligozoospermia. Mol Hum Reprod. 2002;8(3):286–298. | ||
Ferlin A, Tessari A, Ganz F, et al. Association of partial AZFc region deletions with spermatogenic impairment and male infertility. J Med Genet. 2005;42(3):209–213. | ||
Visser L, Westerveld GH, Korver CM, et al. Y chromosome gr/gr deletions are a risk factor for low semen quality. Hum Reprod. 2009;24(10):2667–2673. | ||
Lu C, Zhang J, Li Y, et al. The b2/b3 subdeletion shows higher risk of spermatogenic failure and higher frequency of complete AZFc deletion than the gr/gr subdeletion in a Chinese population. Hum Mol Genet. 2009;18(6):1122–1130. | ||
Zhang S, Tang Q, Wu W, et al. Association between DAZL polymorphisms and susceptibility to male infertility: systematic review with meta-analysis and trial sequential analysis. Sci Rep. 2014;4:4642. | ||
Tüttelmann F, Rajpert-De Meyts E, Nieschlag E, Simoni M. Gene polymorphisms and male infertility–a meta-analysis and literature review. Reprod Biomed Online. 2007;15(6):643–658. | ||
Bartoloni L, Cazzadore C, Ferlin A, Garolla A, Foresta C. Lack of the T54A polymorphism of the DAZL gene in infertile Italian patients. Mol Hum Reprod. 2004;10(8):613–615. | ||
Chen P, Ma M, Li L, et al. Phenotypic expression of partial AZFc deletions is independent of the variations in DAZL and BOULE in a Han population. J Androl. 2010;31(2):163–168. | ||
Fayez AG, El-Sayed AS, El-Desouky MA, et al. Molecular characterization of some genetic factors controlling spermatogenesis in Egyptian patients with male infertility. Int J Infertil Fetal Med. 2012;3(3):69–77. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.