Back to Archived Journals » Reports in Parasitology » Volume 5
Incomplete reversal of genotypic resistance of Plasmodium falciparum to chloroquine after a decade of change in malaria treatment policy in Uganda
Authors Ocan M, Katabazi FA, Kigozi E, Bwanga F, Kyobe S , Ogwal-Okeng J, Obua C
Received 10 December 2015
Accepted for publication 22 January 2016
Published 5 May 2016 Volume 2016:5 Pages 15—22
DOI https://doi.org/10.2147/RIP.S102188
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Manuel Rodriguez Valle
Video abstract presented by Moses Ocan.
Views: 1420
Moses Ocan,1 Fred A Katabazi,2 Edgar Kigozi,2 Freddie Bwanga,2 Samuel Kyobe,2 Jasper Ogwal-Okeng,3 Celestino Obua4
1Department of Pharmacology and Therapeutics, College of Health Sciences, Makerere University, Kampala, Uganda; 2Department of Medical Microbiology, College of Health Sciences, Makerere University, Kampala, Uganda; 3Office of the Vice Chancellor, Lira University, Lira, Uganda; 4Office of the Vice Chancellor, Mbarara University of Science and Technology, Mbarara, Uganda
Background: The potential re-emergence of Plasmodium falciparum parasites sensitive to chloroquine provides an opportunity for the reintroduction of the drug in patient care. With the recent discovery and spread of artemisinin resistance in South east Asia, the emergence of chloroquine sensitivity gives hope for malaria treatment globally. In this study, we explored the prevalence of genotypic chloroquine resistant P. falciparum isolates collected from symptomatic patients in northern Uganda.
Methods: Finger-prick capillary blood spotted on Whatman 903 filter papers were collected from adult symptomatic outpatients (≥18 years) in Lira and Gulu regional referral hospitals. Patients were screened for the presence of Plasmodium infection using rapid diagnostic test histidine rich protein-2 (HRP- 2) prior to blood sample collection. Parasite DNA was extracted from individual spots on the filter papers using chelex-resin method. Presence of mutations, Pfcrt K76T and Pfmdr N86Y were analyzed using polymerase chain reaction-restriction fragment length polymorphism (RFLP) method. A total of 213 and 169 amplicons were analyzed for Pfcrt K76T and Pfmdr N86Y polymorphisms, respectively. The data were analyzed in an Excel 2007 spreadsheet.
Results: A total of 89/213 (41.8%) of the P. falciparum isolates analyzed for Pfcrt K76T polymorphism had wild type (chloroquine sensitive) genotype (76K). The majority, 116/213 (54.4%) carried the mutant (chloroquine resistant) genotype 76T whereas 8/213 (3.8%) had mixed genotypes Pfcrt-76K/T (sensitive/resistant). For Pfmdr N86Y polymorphisms, the majority, 164/169 (97%) of the isolates had wild type (chloroquine sensitive) genotype Pfmdr 86N. A small proportion, (3/169) 1.8% had mutant (chloroquine resistant) genotype Pfmdr 86Y, whereas 2/169 (1.2%) samples had mixed genotypes Pfmdr1-86N/Y (sensitive/resistant).
Conclusion: P. falciparum parasites with genotypic resistance to chloroquine have persisted in the population after more than a decade since the change of policy in Uganda.
Keywords: wild type, resistant, northern Uganda
Background
Malaria remains a disease of public health concern in Uganda, with Plasmodium falciparum parasite responsible for most of the cases.1 The disease causes high mortality especially among children <5 years and expectant mothers. Chloroquine (CQ) was an efficacious, safe, and affordable antimalarial agent that formed the cornerstone of malaria treatment globally in the 1950s and 1960s.2 However, its use was compromised by the development and worldwide spread of resistance. CQ resistance reached alarming levels in most malaria endemic areas of the world.3 In Uganda, for example, CQ resistance peaked at 100% according to previous studies.2,4 Subsequently, the World Health Organization recommended a change in the malaria treatment policy to the current artemisinin-based combination therapy (ACT) globally. In Uganda, national policy for treatment of uncomplicated malaria was first changed from CQ monotherapy to CQ plus sulfadoxine-pyrimethamine combination therapy in 2000. However, this was also soon changed to the current ACT antimalarial agents in 2004 due to resistance.5
Artemisinin agents are highly efficacious with rapid parasite clearance rates and thus offer rapid resolution of malaria symptoms. However, unlike CQ, artemisinin derivatives are more expensive in addition to being difficult to take, especially due to the high pill burden. In addition, reports are emerging of delayed parasite clearance and potential occurrence of resistance to artemisinin antimalarial agents in Cambodia.6 Based on prior experience with CQ where resistance first emerged from Asia and subsequently spread to other parts of the world,3 the current discovery of artemisinin resistance in Cambodia presents a threat to malaria treatment and control programs globally. 7
The antimalarial action of CQ involves interference of hematin detoxification in parasites’ digestive vacuole.8 CQ resistant plasmodium parasites have a decreased ability to concentrate the drug inside the digestive vacuole due to mutations in the Pfcrt K76T gene (threonine to lysine) encoding the drug transporter located on the membrane of the parasite’s digestive vacuole.9 Furthermore, polymorphisms in codon 86 (asparagine to tyrosine, 86Y) of Pfmdr1 gene that encodes P-gh1 have a modulatory role in CQ resistance among parasites that have 76T mutation.10 This mutation (86Y) has been shown to alter transport activity of P-gp required in concentrating CQ inside parasites’ digestive vacuole.11 Exposure to antimalarial agents selects for mutations that allow parasites to survive and continue replicating in the presence of the drug.12 Parasite resistance confers a fitness cost, thus removal of drug pressure potentially leads to reversal to the fit wild type genotype.12,13 This reversal can occur through back mutation or selection of wild type parasites which survive in the presence of the drug.14 In Malawi, Plasmodium parasites with genotypic sensitivity to CQ have re-emerged following long periods of its withdrawal from use in malaria treatment.15 This has been shown to be due to re-expansion of wild type plasmodium parasites.16
However, unlike in Malawi where CQ use was stopped, in Uganda the drug continues to be used in patient care despite its withdrawal following the change in malaria treatment policy.17 For example, according to the Uganda National Clinical Guidelines, CQ is still recommended for use in malaria prophylaxis among sickle cell anemic patients.18 The drug can also be accessed over-the-counter from private drug outlets such as drug stores.19 The continued use of CQ maintains drug pressure in the population and could have a bearing on re-emergence of susceptible P. falciparum parasites in northern Uganda as demonstrated by previous studies in Malawi,15 Kenya,20 and Ethiopia.21 Therefore, the current study was intended to assess the prevalence of genotypic resistance markers for CQ (Pfcrt K76K and Pfmdr1 N86Y) in northern Uganda, an area where CQ can still be accessed and used without a prescription in communities despite malaria treatment policy change.19
Methods
Study design, study area
This was a cross-sectional study and field data collection was done in Lira and Gulu regional referral hospitals in northern Uganda. Northern Uganda is an area of high malaria transmission in the country. Lira and Gulu are ~300 km from the capital Kampala. Lira regional referral hospital is a 415-bed-capacity public hospital serving a population of over 2.5 million. The hospital receives ~131,296 patients annually in the general outpatient department. Gulu regional referral hospital is a 397-bed-capacity public hospital serving a population of over 1.5 million people. The hospital receives ~77,128 patients annually in the general outpatient department.
Study population and sampling procedure
This study was conducted among adult (≥18 years) outpatients presenting to the general outpatient departments of Lira and Gulu regional referral hospitals from August 2013 to May 2014. Patients waiting to see the doctor were selected following systematic random sampling method using intervals of four and three in Lira and Gulu regional referral hospitals, respectively. Those who provided written consent to participate in the study were then screened for presence of Plasmodium infection using rapid diagnostic test (RDT) (HRP-2). Finger-prick capillary blood was then collected and spotted on Whatman 903 filter papers from those patients who were positive for Plasmodium infection using RDT test.
Parasite isolates
Plasmodium parasite infection was diagnosed in the laboratory using RDT. Capillary blood samples were screened for presence of HRP-2, a soluble protein which is expressed only by P. falciparum trophozoites among malaria parasites using RDT-HRP-2. The patients whose blood samples were positive for Plasmodium infection were notified and more blood was requested for further analysis using polymerase chain reaction (PCR). From those who provided written informed consent, more capillary blood from a finger-prick was obtained after sterilizing the fingertip using alcohol swabs. The first drop of blood was cleaned away and the subsequent 2–3 drops were collected on four blood spots on the filter paper (Whatman 903). The blood blotted filter paper was left to dry at room temperature prior to storage at 4°C awaiting PCR analysis. This procedure was repeated for all the recruited patients. A total of 215 blood spotted Whatman No 903 filter papers were collected. The filter papers were then transported to the Molecular Diagnostics laboratory in the Department of Medical Microbiology Makerere University, Kampala, Uganda for PCR analysis.
Plasmodium DNA extraction
Parasite DNA was extracted from dried blood spots on the filter paper using chelex method as previously described by Plowe et al.22 Briefly, two 6 mm blood blotted filter paper discs were cut, each from a separate blood spot on the same filter paper using a sterile single hole punching machine. The discs were soaked in phosphate-buffered saline (PBS) (1 mL) and 10% saponin (50 μL) in a microfuge tube, inverted several times and stored at 4°C overnight. The microfuge tubes were then centrifuged at 13,000 rpm for 5 seconds and the reddish supernatant aspirated. After discarding the supernatant, 1 mL of PBS (no saponin) was added, the tubes inverted several times, and incubated at 4°C for 30 minutes. The microfuge tubes were then centrifuged at 13,000 rpm for 2 minutes and all the saponin and PBS aspirated. Fifty microliters of 10% chelex was then added into the microfuge tube containing the filter paper discs. To each tube 100 μL of sterile water was added. Parasite DNA was extracted by incubating the tubes for 10 minutes in a 95°C heat-block while vigorously vortexing each tube every 2 minutes throughout the incubation period. After incubation the tubes were centrifuged for 5 minutes at 13,000 rpm, then the solution was transferred to separate microfuge tubes. These were then centrifuged at 13,000 rpm for 10 minutes and the final white-to-yellowish supernatant was transferred to separate labeled tubes taking care not to pipette the chelex. The tubes were then stored at −20°C until analysis.
Detection of 76T polymorphism in Pfcrt gene using nested and allele-restricted PCR
Polymorphisms (K76T) in the Pfcrt gene were assessed using a nested PCR amplification followed by restriction fragment length polymorphism (RFLP) method as previously described by Fidock et al.23 The following primers were used in the primary PCR, CRT1F-5′-GACGAGCGTTATAGAGAATTA-3′ and CRT1R-5′-CCAGTAGTTCTTGTAAGACC-3′ to amplify the region flanking codon 76. In the second round PCR (nested), the following primers, CQRAF-5′-TGTGCTCATGTGTTTAAACTT-3′ and CQRBR-5′- CAAAACTATAGTTACCAATTTTG-3′ were used to amplify the PCR products of the primary reaction. In the primary PCR, 2 μL of template DNA was amplified in a 50 μL reaction volume using the following cycling protocol: 95°C for 5 minutes for initial denaturation, 35 cycles of 94°C for 30 seconds, 47°C for 45 seconds, and 68°C for 90 seconds, and a final extension of 68°C for 10 seconds. For the nested PCR, 2 μL of primary PCR products were amplified under the same conditions except for the primers used.
Detection of 86Y polymorphism in Pfmdr1 gene using nested and allele-restricted PCR
The polymorphisms (N86Y) in the Pfmdr1 gene were determined using PCR-RFLP method as previously described by Djimde et al.24 The following primers were used in the primary PCR, Pfmdr1-FOut-5′- TGTTGAAAGATGGGTAAAGAGCAGAAAGAG-3′ and Pfmdr1-ROut -5′-TACTTTCTTATTACATATGACACCACAAACA-3′ to amplify the region flanking codon 86. In the second round, PCR (nested) the following primers, Pfmdr1-FIner-5′- AAAGATGGTAACCTCAGTATCAA AGAAGAG-3′ and Pfmdr1-RIner-5′-GTCAAACGTGCATTTTTTATTAATGACCATTTA-3′ were used to amplify the PCR products of the primary reaction. Two microliters of template DNA was amplified in a 50 μL reaction volume under the following cycling conditions: 94°C for 5 minutes for initial denaturation, 40 cycles of 94°C for 30 seconds, 45°C for 45 seconds, and 68°C for 90 seconds, and a final extension of 68°C for 10 seconds. In the nested PCR, 2 μL of primary PCR products were amplified using the same conditions except for the primers used.
Quality control
Genomic DNA from P. falciparum clones 3D7 (CQ sensitive) and Dd2 (CQ resistant) were used as positive controls and uninfected spots on the filter paper were extracted (free of blood) as negative controls. The nested PCR products were evaluated to check for amplification before digestion by restriction enzymes. This was done by electrophoresis using 2% agarose stained with ethidium bromide and viewed under ultraviolet (UV) trans-illuminator. After running all the DNA samples, two samples per run were randomly selected and re-analyzed using the same conditions to check for consistency of the results.
Restriction digestion with APO I and Afl III
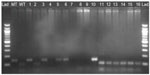
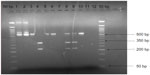
The final amplified nested products were digested with APO I for the Pfcrt K76T and Afl III for Pfmdr N86Y polymorphisms (New England Biolabs, Ipswich, MA, USA). In the reactions, 10 μL of the nested PCR product reaction mixture was treated directly with 2 μL of the restriction enzyme by incubating at 50°C for 6 hours as recommended by the manufacturer (New England Biolabs). The enzyme APO I recognizes and cuts products with wild type 76K genotype releasing two fragments (100 and 49 bp). However, it does not cut the products containing mutant 76T genotype (hence only one band, 149 bp) (Figure 1). The enzyme Afl III recognizes and cuts products containing mutant 86Y genotype releasing two fragments (328 and 232 bp). The enzyme does not cut products containing wild type 86N genotype (thus only one fragment, 560 bp) (Figure 2). The digested products were separated using electrophoresis on 2% agarose gel, stained with ethidium bromide, and the results viewed using UV light in a trans-illuminator.
Data management and analysis
Laboratory data were entered and analyzed in an Excel 2007 spreadsheet. The main outcome variable was presence of Pfcrt K76T and Pfmdr N86Y mutations in parasite DNA samples. The proportion of malaria parasites which carried this mutation was established. Two samples from each of the different PCR runs were randomly selected and re-analyzed to validate the reproducibility of the experiment.
Ethical clearance
The study protocol was reviewed and approved by the School of Medicine Makerere University research and ethics review committee (protocol number: REC REF 2012-072) and Uganda National Council for Science and Technology (protocol number: HS 126). Permission to conduct the study was also obtained from district and hospital authorities. Written informed consent was obtained from all participants who agreed to take part in the study.
Results
Prevalence of Pfcrt K76T polymorphisms in northern Uganda
Amplification of Pfcrt was successful in 213 of the 215 parasite DNA samples analyzed. The majority, 116 (54.4%) of the amplicons contained 76T mutation which is indicative of genotypic CQ resistance. Over a third of the samples, 89 (41.8%) contained K76 genotype which shows genotypic susceptibility to CQ. A proportion, 3.8% (8/213) of the samples contained mixed genotypes (76K/T) which are both CQ resistant and susceptible parasite strains present in the same individual (Table 1).
Prevalence of Pfmdr N86Y polymorphisms in northern Uganda
  | Table 1 Pfmdr1-N86Y and Pfcrt-K76T polymorphisms in the Plasmodium falciparum isolates |
Amplification of Pfmdr1 was successful in 169 of the 215 parasite DNA samples analyzed. The majority, 97% (164/169) of parasite DNA samples contained wild type N86 genotype of the codon Pfmdr1-86 (CQ sensitive genotype). Three samples, 1.8% (3/169) had a mutation 86Y in the Pfmdr1-86 codon (CQ resistant genotype). Two samples, 1.2% (2/169) contained mixed parasite genotypes (86N/Y) which are both wild type and mutant strains (Table 1). We found that the majority (95.5%) of the parasites which had the Pfcrt K76T mutation did not simultaneously carry the Pfmdr N86Y polymorphism.
Discussion
With no effective vaccine against malaria, use of drugs remain the only management currently available despite the challenge of high risk of resistance development. However, withdrawing one medicine from use due to resistance development has been shown to remove drug pressure and potentially lead to the reversal of parasite resistance.15
In this study we found a high prevalence of wild type Pfmdr1-86N genotype (>95%), a result similar to that of a previous study.25 This could be due to the widespread use of ACTs in Uganda as it is the first-line drug in the treatment of uncomplicated malaria.18,26 Studies have shown that wild type Pfmdr1-86N parasite genotype is selected in the population by exposure to ACT antimalarial agents.27,28 A higher prevalence of wild type Pfmdr-86N genotype was observed in Kenya after the roll out of ACT use.20 In addition, pooled analysis of data from multiple clinical trials indicated a positive relation between high prevalence of wild type Pfmdr-86N genotype and use of artemether-lumefantrine antimalarial agents.29 In our study, the prevalence of wild type Pfmdr-86N genotype was higher than that of Pfcrt-76K following the introduction of ACTs in malaria treatment over a decade ago. This finding is consistent with that of previous studies30,31 where emergence of wild type 86N genotype preceded that of 76K after implementation of ACT use. Although artemether-lumefantrine is still very effective in malaria treatment in Uganda,32 analysis of data from different clinical trials showed that high prevalence of wild type Pfmdr-86N parasite genotype is associated with decreased parasite sensitivity to artemisinin-lumefantrine.29
Furthermore, we found higher prevalence (41.8%) of wild type 76K P. falciparum genotype than a previous study by Mbogo et al33 that reported 17% in Tororo, Uganda. The variation in the findings could be due to the difference in the study periods. In the study by Mbogo et al,33 samples of previous clinical trials collected between 2002 and 2012 were used in the analysis, which included samples collected when CQ was still a recommended drug in the CQ sulfadoxine-pyrimethamine combination, unlike samples collected in 2013 to 2014 used in the current study. The findings of this study in addition to that of Mbogo et al,33 are however indicative of a steady decline in the prevalence of mutant 76T alleles of the Pfcrt-76 gene in Uganda which had previously peaked at 100% in the population. Studies have shown that it took approximately 8 years (1993 to 2001) since the change in malaria treatment policy for CQ sensitivity to re-emerge in Malawi.15 However, in Uganda during the same amount of time (10 years; 2005 to 2015), the prevalence of CQ sensitive P. falciparum genotypes is still lower than that observed in Malawi as shown by our findings.15
Reversal of CQ resistance seems to follow complete removal of drug pressure in the population.13,15 In Malawi, for example, following the change in malaria treatment policy, CQ importation into the country was stopped. This had a remarkable effect in removing drug pressure, hence the re-emergence of CQ susceptible plasmodium parasites in the population.15 In Uganda unlike Malawi, even after the change in malaria treatment policy,5 CQ importation and use was not completely eliminated. Studies have reported low levels of continued CQ use in Uganda.19,34 In addition, CQ is still recommended for use in rheumatoid arthritis and malaria prophylaxis among sickle cell patients in Uganda.18 In Ghana, the risk of infection with Plasmodium falciparum parasites carrying Pfcrt 76T mutation (CQ resistant) was higher in areas where malaria was treated by using CQ.35 Therefore, the continued use of CQ in the population could explain the low rate of reversal in CQ resistance found in the current study.36,37 In addition, the potential development of compensatory mutations among P. falciparum parasites could also play a role in the persistence of the resistant Pfcrt-76T mutations in the population.12,38,39 Unlike Malawi, where CQ resistance remained at ~85% prior to the change in malaria treatment policy in 1993,36 in Uganda, the prevalence of CQ resistance had peaked at 100% prior to the change in malaria treatment policy in 2005.4 This variation in the extent of CQ resistance prior to change in malaria treatment policy may explain the slow reversal in CQ resistance in Uganda as compared to Malawi, as shown in the findings of this study. Some patient samples had mixed Pfcrt-76K/T genotypes. A previous study by Mita et al40 showed that patients with mixed infections had sensitive in vivo CQ response.
Studies done in other African countries show different prevalence rates of CQ resistant Pfcrt 76T gene following change in malaria treatment policy from CQ to artemisinin. In Ghana, for example, the prevalence of Pfcrt T76 declined from 80% to ~60% between 2005 and 2011.41 A study by Mohammed et al42 in Tanzania found a CQ susceptibility rate of more than 90% among P. falciparum parasite isolates following 12 years after the change in policy. In Kenya, when CQ use in malaria treatment was stopped from 1993 to 2006, CQ susceptibility rose from 5% to 40%. Similarly, in Mozambique, CQ susceptibility rose from 5% to 80% within 5 years of policy change,43 while in Malawi there was 100% CQ susceptibility recorded in 13 years following the change in malaria treatment policy.15,36 Thus, there is evidence that the change in malaria treatment policy has had an effect on reversal of CQ susceptibility in different African countries. However, the variation in the rate of recovery of CQ susceptibility across several African countries is likely due to the lack of synchronized implementation of the policy. In addition, other factors such as the variation in the intensity of malaria transmission in different geographical locations coupled with the introduction of closely related antimalarial agents such as amodiaquine may further explain the observed variation.42
The current study provides evidence of a slow progressive decline in the prevalence of genotypic (Pfcr K76T and Pfmdr N86Y) CQ resistance among P. falciparum parasites in Uganda. This is indicative of the persistent possibility of unfavorable clinical outcomes if CQ is introduced in malaria treatment in the country. However, Chaijaroenkul et al, Francis et al, and Dorsey et al4,44,45 in their studies showed that in high transmission areas like northern Uganda,46 clinical response to treatment of resistant parasites using CQ may also be influenced by other factors such as immunity.
There was unsuccessful amplification in some of the samples, especially for the Pfmdr1 codon that is similar to the findings of other studies.29 This could have been due to low concentration of template (parasite) DNA, which is likely as we did not quantify parasite load during field sample collection.
Conclusion and recommendation
The current finding indicates presence of partial reversal of genotypic CQ resistance of Pfcrt K76T in northern Uganda following a decade of incomplete withdrawal from use in the country. Continued use of CQ in hospitals and communities, albeit at low levels, could have a bearing on the persistence of CQ resistance genotype in the population. Therefore, complete removal of drug pressure through enforcement of regulations, especially on, importation and sale of CQ, is needed. There is a need for continuous surveillance of Pfmdr1 and Pfcrt plasmodium parasite polymorphisms in the country to help policy makers manage the challenge of drug resistance in malaria treatment.
Author contributions
MO, CO, JOO, and FB conceptualized and designed the study. MO did the sample collection and prepared the draft manuscript. FAK, EK, SK, and MO performed the molecular analysis of the work. KF did the genotyping of the results. All authors contributed to the interpretation of data, writing and revising the manuscript, and have approved the final version.
Acknowledgments
This study was funded by grant number 5R24TW008886 supported by the Office of the US Global Aids Coordinator, the National Institute of Health, and the Health Resources and Services Administration. This work obtained additional funding from the Swedish Research Council VR-link award, grant number VR2011-7381, and the African Regional Capacity Development for Health Services and Systems Research (ARCADE-HSSR), FP7 grant agreement number 265970.
The authors sincerely thank the laboratory technicians at Lira and Gulu regional referral hospitals and the study participants.
Disclosure
The authors report no conflicts of interest in this work.
References
Yeka A, Gasasira A, Mpimbaza A, et al. Malaria in Uganda: challenges to control on the long road to elimination: I. Epidemiology and current control efforts. Acta Trop. 2012;121(3):184–195. | |
Frosch AE, Venkatesan M, Laufer MK. Patterns of chloroquine use and resistance in sub-Saharan Africa: a systematic review of household survey and molecular data. Malar J. 2011;10:116. | |
Naidoo I, Roper C. Mapping ‘partially resistant’, ‘fully resistant’, and ‘super resistant’ malaria. Trends Parasitol. 2013;29(10):505–515. | |
Dorsey G, Kamya MR, Singh A, Rosenthal PJ. Polymorphisms in the Plasmodium falciparum pfcrt and pfmdr-1 genes and clinical response to chloroquine in Kampala, Uganda. J Infect Dis. 2001;183(9):1417–1420. | |
Nanyunja M, Nabyonga Orem J, Kato F, Kaggwa M, Katureebe C, Saweka J. Malaria treatment policy change and implementation: the case of Uganda. Malar Res Treat. 2011;2011:683167. | |
Ariey F, Witkowski B, Amaratunga C, et al. A molecular marker of artemisinin resistant Plasmodium falciparum malaria. Nature. 2014; 505(7481):50–55. | |
World Health Organization. Community-Based Surveillance of Antimicrobial Use and Resistance in Resource-Constrained Settings. Geneva: World Health Organization; 2009. Available from: http://apps.who.int/medicinedocs/documents/s16168e/s16168e.pdf. Accessed March 6, 2016. | |
Dorn A, Stoffel R, Matile H, Bubendorf A, Ridley RG. Malarial haemozoin/beta-haematin supports haem polymerization in the absence of protein. Nature. 1995;374(6519):269–271. | |
Sidhu AB, Verdier-Pinard D, Fidock DA. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science. 2002;298(5591):210–213. | |
Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403(6772):906–909. | |
Sanchez CP, Stein W, Lanzer M. Trans stimulation provides evidence for a drug efflux carrier as the mechanism of chloroquine resistance in Plasmodium falciparum. Biochemistry. 2003;42(31):9383–9394. | |
Levin BR, Perrot V, Walker N. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics. 2000;154(3):985–997. | |
Austin DJ, Kristinnson KG, Anderson RM. The relationship between the volume of antimicrobial consumption in human communities and frequency of resistance. Proc Natl Acad Sci U S A. 1999;96(3):1152–1156. | |
Seppala H, Klaukka T, Lehtonen R, Nenonen E, Huovinen P. Outpatient use of erythromycin: link to increased erythromycin resistance in group A streptococci. Clin Infect Dis. 1995;21(6):1378–1385. | |
Laufer MK, Thesing PC, Eddington ND. Return of chloroquine antimalarial efficacy in Malawi. N Engl J Med. 2006;355(19):1959–1966. | |
Laufer MK, Takala-Harrison S, Dzinjalamala FK, Stine OC, Taylor TE, Plowe CV. Return of chloroquine-susceptible falciparum malaria in Malawi was a reexpansion of diverse susceptible parasites. J Infect Dis. 2010;202(5):801–808. | |
Nakibuuka V, Ndeezi G, Nakiboneka D, Ndugwa CM, Tumwine JK. Presumptive treatment with sulphadoxine-pyrimethamine versus weekly chloroquine for malaria prophylaxis in children with sickle cell anemia in Uganda: a randomized controlled trial. Malar J. 2009;8:237. | |
Uganda Clinical Guidelines. National Guidelines for Management of Common Conditions. Kampala: Ministry of Health; 2012. Available from: http://apps.who.int/medicinedocs/documents/s21741en/s21741en.pdf. Accessed April 12, 2016. | |
Ocan M, Bwanga F, Bbosa GS, et al. Patterns and predictors of self-medication in northern Uganda. PLoS ONE. 2014;9(3):e92323. | |
Mwai L, Ochong E, Abdirahman A, et al. Chloroquine resistance before and after its withdrawal in Kenya. Malar J. 2009;8:106. | |
Mekonnen SK, Aseffa A, Berhe N, et al. Return of chloroquine-sensitive Plasmodium falciparum parasites and emergence of chloroquine-resistant Plasmodium vivax in Ethiopia. Malar J. 2014;13:244. | |
Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg. 1995;52(6):565–568. | |
Fidock DA, Nomura T, Talley AK, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6(4):861–871. | |
Djimde A, Doumbo OK, Cortese JF, et al. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med. 2001;344(4):257–263. | |
Lekana-Douki JB, Boutamba SDD, Zatra R, et al. Increased prevalence of the Plasmodium falciparum Pfmdr1 86N genotype among field isolates from Franceville, Gabon after replacement of chloroquine by artemether-lumefantrine and artesunate-mefloquine. Infect Genet Evol. 2011;11(2):512–517. | |
World Health Organization. Guidelines for the Treatment of Malaria. Geneva: World Health Organization; 2010. Available from: http://apps.who.int/iris/bitstream/10665/162441/1/9789241549127_eng.pdf. Accessed March 3, 2016. | |
Sisowath C, Stromberg J, Martensson A, et al. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem). J Infect Dis. 2005;191(6):1014–1017. | |
Duraisingh MT, Triglia T, Ralph SA, et al. Phenotypic variation of Plasmodium falciparum merozoite proteins directs receptor targeting for invasion of human erythrocytes. EMBO J. 2003;22(5):1047–1057. | |
Venkatesan M, Gadalla NB, Stepniewska K, et al. Polymorphisms in Plasmodium falciparum chloroquine resistance transporter and multidrug resistance 1 genes: parasite risk factors that affect treatment outcomes for P. falciparum malaria after artemether-lumefantrine and artesunate-amodiaquine. Am J Trop Med Hyg. 2014;91(4):833–843. | |
Sisowath C, Petersen I, Veiga MI, et al. In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether-lumefantrine in Africa. J Infect Dis. 2009;199(5):750–757. | |
Yang Z, Zhang Z, Sun X, et al. Molecular analysis of chloroquine resistance in Plasmodium falciparum in Yunnan Province, China. Trop Med Int Health. 2007;12(9):1051–1060. | |
Muhindo MK, Kakuru A, Jagannathan P, et al. Early parasite clearance following artemisinin-based combination therapy among Ugandan children with uncomplicated Plasmodium falciparum malaria. Malar J. 2014;13:32. | |
Mbogo GW, Nankoberanyi S, Tukwasibwe S, et al. Temporal changes in prevalence of molecular markers mediating antimalarial drug resistance in a high Malaria transmission setting in Uganda. Am J Trop Med Hyg. 2014;91(1):54–61. | |
Ministry of Health (MOH). Access to and Use of Medicines by Households in Uganda. Kampala: Ministry of Health; 2008. Available from http://apps.who.int/medicinedocs/documents/s16374e/s16374e.pdf. Accessed March 2, 2016. | |
Asare KK, Boampong JN, Afoakwah R, Ameyaw EO, Sehgal R, Quashie NB. Use of proscribed chloroquine is associated with an increased risk of pfcrt T76 mutation in some parts of Ghana. Malar J. 2014; 13:246. | |
Kublin JG, Cortese JF, Njunju EM, et al. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis. 2003;187(12):1870–1875. | |
Mayor AG, Gomez-Olive X, Aponte JJ, et al. Prevalence of the K76T mutation in the putative Plasmodium falciparum chloroquine resistance transporter (pfcrt) gene and its relation to chloroquine resistance in Mozambique. J Infect Dis. 2001;183(9):1413–1416. | |
Sherman DR, Mdluli K, Hickey MJ, et al. Compensatory ahpC gene expression in isoniazid-resistant Mycobacterium tuberculosis. Science. 1996;272(5268):1641–1643. | |
Pelleau S, Moss EL, Dhingra SK, et al. Adaptive evolution of malaria parasites in French Guiana: reversal of chloroquine resistance by acquisition of a mutation in pfcrt. Proc Natl Acad Sci U S A. 2015;112(37):11672–11677. | |
Mita T, Venkatesan M, Ohashi J, et al. Limited geographic origin and global spread of sulfadoxineresistant dhps alleles in Plasmodium falciparum populations. J Infect Dis. 2011;204(12):1980–1988. | |
Afoakwah R, Boampong JN, Egyir-Yawson A, Nwaefuna EK, Verner ON, Asare KK. High prevalence of PfCRT K76T mutation in Plasmodium falciparum isolates in Ghana. Acta Trop. 2014;136:32–36. | |
Mohammed A, Ndaro A, Kalinga A, et al. Trends in chloroquine resistance marker, Pfcrt-K76T mutation ten years after chloroquine withdrawal in Tanzania. Malar J. 2013;12:415. | |
Thomsen TT, Madsen LB, Hansson HH, et al. Rapid selection of Plasmodium falciparum chloroquine resistance transporter gene and multidrug resistance gene-1 haplotypes associated with past chloroquine and present artemether-lumefantrine use in Inhambane District, southern Mozambique. Am J Trop Med Hyg. 2013;88(3):536–541. | |
Francis D, Nsobya SL, Talisuna A, et al. Geographic differences in antimalarial drug efficacy in Uganda are explained by differences in endemicity and not by known molecular markers of drug resistance. J Infect Dis. 2006;193(7):978–986. | |
Chaijaroenkul W, Ward SA, Mungthin M, et al. Sequence and gene expression of chloroquine resistance transporter (pfcrt) in the association of in vitro drugs resistance of Plasmodium falciparum. Malar J. 2011;10:42. | |
Proietti C, Pettinato DD, Kanoi BN, et al. Continuing intense malaria transmission in northern Uganda. Am J Trop Med Hyg. 2011;84(5):830–837. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.