Back to Journals » International Journal of Nanomedicine » Volume 14
In-vitro Detection of Phytopathogenic Fungal Cell Wall by Polyclonal Sera Raised Against Trimethyl Chitosan Nanoparticles
Authors Joshi H, Malik A , Aggarwal S , Munde M, Maitra SS, Adlakha N, Bhatnagar R
Received 23 June 2019
Accepted for publication 3 September 2019
Published 20 December 2019 Volume 2019:14 Pages 10023—10033
DOI https://doi.org/10.2147/IJN.S220488
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Thomas Webster
Hemant Joshi,1,* Anshu Malik,1,* Soumya Aggarwal,2 Manoj Munde,2 Subhrangsu Sundar Maitra,1 Nidhi Adlakha,3 Rakesh Bhatnagar1,4
1Molecular Biology and Genetic Engineering Laboratory, School of Biotechnology, Jawaharlal Nehru University, New Delhi 110067, India; 2School of Physical Science, Jawaharlal Nehru University, New Delhi 110067, India; 3Regional Centre for Biotechnology, NCR Biotech Cluster, Faridabad 121001, India; 4Banaras Hindu University, Banaras, Uttar Pradesh 221005, India
*These authors contributed equally to this work
Correspondence: Rakesh Bhatnagar
Molecular Biology and Genetic Engineering Laboratory, School of Biotechnology, Jawaharlal Nehru University, New Delhi 110067, India
Email [email protected]
Nidhi Adlakha
Regional Centre for Biotechnology, NCR Biotech Cluster, Faridabad 121001, India
Email [email protected]
Purpose: The objective of this research was to generate a tool for the first-line detection of fungal infection in plants. Chitin is one of the unique fungal cell wall polysaccharide which is naturally deacetylated to chitosan upon infection. It is said to be involved in the fungal cell wall modulation and plant-pathogen communication. Therefore, detection of chitosan could be potentially helpful in the detection of fungal contamination.
Methods: Five different phytopathogenic fungi strains were used for the study. Polyclonal sera were raised in the mice against Trimethylchitosan nanoparticles to generate an enhanced humoral immune response and generate a rich and heterogeneous repertoire of antibodies. The binding affinity of the sera with fungal cell wall was analyzed by ELISA, Langmuir isotherm, confocal microscopy and ITC (Isothermal Calorimetry).
Results: The raised polyclonal sera could detect chitosan in the fungal cell wall, as analyzed with the different techniques. However, the detection specificity varied among the strains in proportion to the chitin content of their cell wall. Fusarium oxysporum was detected with the highest affinity while Trichoderma reesei was detected with the least affinity by ELISA. Adsorption isotherm, as well as ITC, revealed the specific and high binding capacity. Confocal microscopy also confirmed the detection of all strains used in the study.
Conclusion: This novel technique employing TMC nanoparticulate system could be potentially used as a source to raise sera against chitosan in an inexpensive and less laborious manner. Rapid detection of fungal contamination by the polyclonal antibodies could help in devising a quick solution. The polyclonal sera are expected to detect a span of epitopes and provide precise detection. The detection system could be advanced for future applications such as food quality control, crop protection, and human fungal infection detection and treatment.
Keywords: polyclonal sera, chitosan, chitin, fungus, cell-wall, detection
Introduction
Phytopathogens infect a large number of economically important plants resulting in massive yield loss globally. The effective and early detection strategy is the first measure to control the disease outbreak. Currently, the diagnosis of phytopathogenic fungus relies on traditional staining and culture techniques, however, they face major limitations such as – need to culture the infectious organism, error-prone and time-consuming. Therefore, there is a need for a fast and inexpensive alternative for the detection of pathogenic fungi.
Interestingly, the cell wall of all the fungi is enriched with polysaccharides, which play a pivotal role in a multitude of processes such as growth, morphogenesis and the polysaccharides are even speculated as virulence factors.1 Generally, the cell wall has an outer layer rich in mannosylated proteins and an inner layer essentially encompassing cross-linked chitin-glucan matrix.2 Several phytopathogenic fungal species comprehend more than 7 chitin synthases in their genomes.3 It has been reported that in phytopathogens B. cinerea and U. maydis, elimination of a single chitin synthase lead to an amplified plant immune response due to defects in cell wall conformation.4,5 Ensuing cellulose, chitin is certainly the most abundant polysaccharide in nature. It is universally found in the exoskeletons of crustacean shells and insects as well as in fungal cell walls. Its major functional contribution is providing structural stability and consequently protection to the organism. Chemically it is made up of β-1,4-linked N-acetyl-D-glucosamine (GlcNac) and D-glucosamine (GlcN) residues.6 Nearly hundred billion tons of chitin are produced annually.7 Chitin is insoluble in most aqueous and organic solvents owing to its highly aggregated structure. The hydrogen bonds between the acetyl, amino and hydroxyl groups present in the chain are responsible for its crystalline structure.8 The reactive amino groups present in the chain impart positive charge upon solubilization, which can be chemically increased by increasing the deacetylation of the chain. The Degree of deacetylation (DD) is referred to denote the number of acetyl groups in the chain. Many soluble derivatives of chitin, such as Chitosan, trimethyl chitosan, and many sulphur based derivatives, have been chemically derived and are more soluble than the parent chitin.9 Natural deacetylation of chitin to chitosan is also a process reported to be involved in the fungal cell wall modulation and plant-pathogen communication.10
Upon fungal infection in plants, chitin, and chitosan induce several defense reactions such as reactive oxygen species generation and expression of early responsive and defense-related genes, phytoalexin biosynthesis and callose formation.11–16 The degree of polymerization17,18 and degree of acetylation19,20 determine the scale of response generated in the plants. Several chitinases play a key role in early plant defense response.21 Chitinases provide the nitrogen and carbon by degrading chitin.22 Similarly, chitin deacetylases come into play when fungal hyphae penetrate the plant and chitin is converted to chitosan in order to evade the plant defense mechanisms.23 Chitin to chitosan conversion protects the pathogenic fungi from plant extracellular chitinases because chitosan is a poor substrate for chitinases.20,24 As chitin is not present in prokaryotes and higher plants or animals, its detection can be been employed to detect the fungal contaminations in diseased plants and several food items. As chitosan is more soluble than chitin, it has been used by several groups for developing anti-chitosan antibodies for the detection of fungal pathogens.25–27 These antibodies could be beneficial for quality control purposes in food or other industries. Moreover, plant and animal fungal pathogens can also be detected by utilizing these antibodies. Fusarium-resistant plants have been developed by coupling antifungal antibodies to lytic enzymes.28,29 Trimethylchitosan (TMC), the quaternized form of chitosan, offers many applications as it is biodegradable, non-toxic to animals, soluble in aqueous medium and is much more tractable than chitosan.30,31 Chitosan and TMC have been reported to possess intrinsic adjuvant as well as mucoadhesion characteristics as its nanoparticles can elicit in-vitro T-cell maturation and proliferation.32 Chitosan and chitin have been reported to be antimicrobial in nature. The antimicrobial activity of chitosan against bacteria, yeast, and filamentous fungi has also been studied by various groups.33–37
TMC has garnered a recent interest as a nanoparticulate carrier for vaccine delivery and for generation of heightened immune response.30,31 Furthermore, studies have shown that smaller NPs are easily engulfed by the antigen-presenting cells (APCs) relative to the larger sized NPs and could potentially be used as a drug delivery agent against tumors.38 An enhanced uptake of TMC NPs ranging between 200 and 300nm across the nasal epithelium membrane has been reported owing to their small size.39 Therefore, we report here for the first time, the detection of plant-fungal pathogenic cell wall chitin with the help of this polyclonal sera raised against trimethyl chitosan nanoparticles in mice.
Materials and Methods
Derivation of Trimethyl Chitosan from Chitosan and Mice Immunization
Muzzarelli and Tanfani method with slight modifications was adopted to derive Trimethyl chitosan from chitosan as previously mentioned by Malik et al31,40 In this two-step methylation process, the primary methylation and dissolution of 1.5 g chitosan were carried out with the help of 5 mL formic acid and formaldehyde in 30 mL H2O (Eschweiler-Clarke). For 5 days the solution was heated in reflux conditions at 70°C in an oil bath. After cooling, followed by evaporation, the pH of solution was gradually inflated to 12 by 1NaOH. Dimethylated form of chitosan with a gel-like consistency was obtained, which was further methylated by methyl iodide (0.25 mL) treatment after dissolving it in 12 mL of 1-methyl-2-pyrrolidinone. The reaction was carried out at 40°C for 8 hrs followed by cooling and final product precipitation by 1:1 mixture of diethyl ether and ethanol. The precipitate was isolated by centrifugation and then washed thrice with diethyl-ether. TMC was then dried and dissolved in 25 mL 10%NaCl and dialyzed against distilled water for 3 days. Before an analysis by 1H-nuclear magnetic resonance 1H-NMR spectroscopy for the presence of trimethyl group in chitosan, it was lyophilized to obtain an aqueous soluble dry powder form. The degree of quaternization (DQ) of TMC was calculated from the equation:
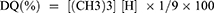
Preparation of TMC Nanoparticles and Characterization
Ionic gelation technique was employed for the preparation of TMC nanoparticles. A 2 mg/mL solution of TMC in 10 mM HEPES buffer was treated with 1 mL Tripolyphosphate (TPP) aqueous solution (1.7 mg/mL) drop by drop while continuous stirring. TPP, the crosslinking agent, induces ionic complexation resulting in the formation of an opalescent dispersion indicating the formation of nanoparticles. The nanoparticles were harvested by centrifugation at 15,000 rpm for 10 min on a 10 µL glycerol bed. The particles were stored at −20°C until further use. Particles were resuspended in HEPES buffer pH 7.4 and after a brief sonication, the size, polydispersity index, and zeta potential were measured by using Nanosizer ZS (Malvern Instruments, Malvern, UK). Particles were observed and analyzed by SEM and TEM imaging. For imaging sample preparation the particles were processed as described earlier by Malik et al.31
Fifty micrograms of aqueous soluble TMC nanoparticles were subcutaneously injected individually to 6–8-week old female Balb/c mice (n=8) with or without CpG ODN (20 µg/mice). A booster dose was given 2 weeks post-immunization and mice sera were collected 28 days after the immunization. The pooled sera were stored at −80°C until further use.
Ethical Statement
All mice experiments were conducted in agreement with Institutional Animal Ethics Committee (Jawaharlal Nehru University) and Council for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA, Ministry of Social Justice and Empowerment, Government of India). Mice were kept in the individually ventilated animal caging system.
Plant Pathogenic Fungus Employed for the Study
Pure fungal isolates of Penicillium funiculosum ITCC7711, Botrytis cinereal ITCC6192, Fusarium oxysporum ITCC3866, Neurospora crassa ITCC524, and Trichoderma reesei ITCC4025 were obtained from Indian Type Culture Collection (ITCC), Indian Agricultural Research Institute, New Delhi, India. All the fungal strains were maintained on Potato dextrose agar (PDA; HiMedia) at 28°C. For spore isolation, the fungal isolates were grown on PDA for 7 days. Spores were scratched out slowly from the surface of agar using a Pasteur pipette and washed three times with sterile PBS by centrifugation (4,000 × g, 10 mins, room temperature). The spores were counted using haemocytometer. For this, 10 μL of spore suspension was pipetted on the corner of the grid and counted with the help of microscope under 40X. Spore density of 108/mL was utilized for further experiments.
Detection of Antibodies Against Chitin
Five hundred spores of all fungal strains were coated on 96 well polystyrene plates overnight at 4°C. Unless otherwise stated, for further reactions, plates were incubated for 1.5 h at room temperature for each step. Microtiter plate was washed with PBS containing 0.05% (v/v) Tween-20 (PBST) thrice after each incubation step. To circumvent any non-specific binding the surface of wells was blocked by incubating with 2% BSA in PBS. In the next step, 100 µL of serial dilutions (1:100 to 1:25,000) of mice sera was incubated with spores. Finally, a secondary 200 ng/mL goat anti-mouse IgG antibody labeled with horseradish peroxidase (HRP) was incubated with spores and 3,3ʹ,5,5ʹ-tetramethylbenzidine (TMB) substrate was used to detect the binding. After 30 min incubation with TMB at room temperature, the reaction was arrested by 1N HCl treatment and the optical density in wells was observed and recorded at 450 nm. The readings were obtained in triplicate and were analyzed to compare the affinity of sera towards the different test spores.
Adsorption Isotherm
The adsorption process is illustrated by the Langmuir model by assuming one molecule adsorbed per adsorption site till a monolayer coverage is achieved, given a specific number of adsorption sites of identical energy.41 The binding isotherm was prepared in order to find out the binding capacity of the polyclonal sera with the Fusarium oxysporum fungal spores. The adsorption coefficient was also calculated from the curve. Ten serial polyclonal antibody dilutions were prepared with 1X PBS (pH 7.4) buffer for each system. Ten-milligram fine chitosan TMC powder was dissolved in 10 mL 1X PBS (pH 7.4). Equal volumes of the antibody working solutions and 1 mg/mL suspensions of chitosan and tri-methyl chitosan were gently mixed in 1.5 mL microcentrifuge tubes by end-over-end rotation at 4°C for 2 hrs. The protein adsorption level was determined by centrifuging the samples at 12,000 g for 15 mins and analyzing the protein content in the supernatant by micro BCA assay. The amount of antibody adsorbed onto chitosan or trimethyl chitosan was determined by subtracting the amount found in the supernatant from the amount added initially.
Immunofluorescence Microscopy/Confocal Based Localization
Immunofluorescence microscopy was implemented to visually examine the binding of the polyclonal sera to the chitin present the fungal cell wall. The round glass coverslips were placed in a 12-well tissue culture plate. To ensure tight coating of spores, 500 µL of the prepared germinated spore solution was spread on the coverslips and the culture plate was centrifuged (1,000 g, 15 mins). After overnight drying, it was treated with 200 µL of 1:1000 dilution of polyclonal sera for 1.5 hrs. Post-incubation spores were washed thrice with PBST. For the bound antibody detection 250 µL (5 µg/mL) of FITC-labeled goat anti-mouse IgG antibody, Sigma-Aldrich, India, was added to the plate and incubated for 1 h. Again, spores were washed thrice with PBST. The inverted coverslips were mounted on glass slides using fluorescence anti-fade agent and sealed for observation under a microscope. Saccharomyces cerevisiae mycelia was used a negative control, as its cell wall does not contain any chitin. A (Olympus FluoviewTm FV1000) confocal microscope was used for viewing and recording the images.
Isothermal Calorimetry Based Affinity Determination: (ITC)
Isothermal titration calorimetry measurements were performed to measure the binding of polyclonal antibody with the Fusarium oxysporum spores at 25°C on a MicroCal iTC200 (Malvern Instruments Ltd. UK). All the antibody and spore fractions were dissolved in the gently degassed 0.1 mM phosphate buffer, pH 7.4. A total volume of 40 µL from injection syringe was added to a sample cell containing 280 µL of spore’s fraction. A total of 20 injections with each injection of 2 µL of polyclonal antibody was titrated into the sample cell containing 500 µg/mL of fungal spore fraction and each injection was separated by 150-s intervals to allow the signal to return to baseline. A constant stirring speed of 300 rpm was maintained to ensure proper mixing after each injection. Control experiments were performed under similar conditions by titrating proteins into the buffer and were subtracted to correct the heat of ligand dilution. Thermodynamic parameters were obtained by fitting the corrected data to non-competitive one sets of sites model using origin software. BSA was used as a negative control.
Results
TMC Derivation and Its Nanoparticles Characterization
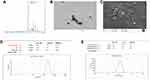
The final product Trimethyl chitosan was obtained by the reductive methylation of chitosan. The quaternized derivative of chitosan was completely aqueous soluble. The D2O soluble TMC was examined for the presence of methyl group. In Figure 1, the H1-NMR profile of the final product reveals the peak of the trimethyl group at the -NH2 group of chitosan at 3.3 ppm. The final product with a degree of quaternization of 84% was utilized for further experiments.
The average particle size was found to be 244.4 ± 12 nm as depicted in Figure 1D. The particles shape and morphology were observed by SEM and TEM as shown in Figure 1B and C. The nanoparticles were found to be smooth in texture and irregular polygonal in shape. The polydispersity index of the particles was found to be 0.16 ± 0.02 indicating fine homogeneity of the particles. Figure 1E reveals the zeta potential of the nanoparticles, i.e. 5.81 ± 0.5 mV which confirmed that the particles carried a positive surface charge.
Characterization of Anti-Chitin Antibodies
After serial dilution titration of sera with the spores, it was established that 1:5000 dilution was optimum for binding. Hence, ELISA based comparative affinity of the polyclonal sera for a particular spore strain was illustrated. At the standardized dilution of sera (1:5000) and fungal spores (500). A differential affinity of an antibody was detected/observed for the spores of different fungal strains. Figure 2 depicts that the Fusarium oxysporum was detected with the highest affinity while Trichoderma reesei was detected with the least affinity. All the strains were detected by the polyclonal antibodies with significantly higher absorbance values than the BSA as well as only spores. The optical density of detection was not significantly different amongst BSA and only spores (not treated with sera). The increasing order of affinity observed by ELISA-based detection was Trichoderma reesei<Penicillium funiculosum<Neurospora crassa<Botrytis cinerea<Fusarium oxysporum. The differential binding could be explained in terms of the varied amount of chitin present in various species of fungi.42
Langmuir Adsorption Isotherm
The Langmuir isotherm was prepared with the Fusarium oxysporum spores exhibiting highest affinity towards the polyclonal sera as observed in ELISA-based detection. A curve 1/qe versus 1/Ce was plotted to investigate the fitting of Langmuir model for the equilibrium data of spore and antibody sorption, from which a straight line was obtained as shown in Figure 3. The correlation coefficient R2 was found to be 0.822 indicating that the data fitted well with the Langmuir model. The spores showed a typical isotherm curve and the Adsorption capacity and the strength of Adsorption were calculated. The adsorption capacity was found to be 2500 mL/mg and the strength of adsorption was 9.16 mL/mg, which demonstrates a robust binding between the polyclonal antibodies and the spores.
 |
Figure 3 Langmuir adsorption isotherm of Fusarium oxysporum spores against polyclonal sera. |
Fungal Cell Wall Detection
The confocal microscopy in Figure 4, revealed positive fluorescence signals on the spore’s surface, confirming the binding and detection of chitin. All the strains were detected by the polyclonal sera. Binding was found to be stronger in case of Neurospora crassa and Botrytis cinerea strains (Figure 4A and B); however, it was relatively weaker in the case of Trichoderma reesei strain (Figure 4C). Negligible traces of fluorescence were detected in case of negative control, Saccharomyces cerevisiae (Figure 4D). The strains which showed least and medium affinity were selected for the immunofluorescence imaging in order to validate the visual detection by the polyclonal sera.
ITC-based Binding of Polyclonal Sera with Fungal Cell Wall
The interactions of antibodies with chitosan and cell wall fraction were characterized using ITC technique. First antibody binding to chitosan Figure 5A1 and cell wall fraction Figure 5B1 data were fitted using two binding sites model, out of two binding sites, the first site binds strongly and specifically as compared to the second binding site in both the cases, as indicated by their K values (for chitosan K1=1.41×109 M−1, K2=1.47×105 M−1 and for cell wall fraction K1=7.19×106 M−1, K2=1.01×104 M−1). In case of second antibody and BSA data were fitted using one set of site model, where it binds strongly to chitosan (Figure 5A(2–3)) as compared to cell wall fraction (Figure 5B(2–3)). In the case of cell wall fraction, binding is so weak or non-specific that it cannot be fitted properly. The binding here is interaction specific because both ΔH and ΔS are negative; indicating that the interactions here are enthalpically driven.
Discussion
Polyclonal antibody-based detection of chitin in pathogenic fungal cell wall has been previously reported in several studies.43,44 In previous reports, polyclonal antibodies have been raised against chitosan by conjugating it with BSA and hemocyanin carrier proteins or by haptenizing chitosan combined with several booster dosages in animals to achieve an optimal humoral response.25,26,45 Our technique of generating antibody repertoire bears the benefit of a single booster dose as well as employment of adjuvant properties of TMC and its nanoparticles. Polyclonal sera are also able to span more epitopes than any monoclonal antibody. Chitosan’s adjuvant properties have been in exploration for quite some time. There are reports suggesting its immunogenic response in mice models as well its heightened response when used in combination with any antigenic carrier protein. Its FDA approval for tissue engineering and wound healing makes it interesting biodegradable material to be explored for its future applications.46 Chitosan matrix based encapsulation of bioactives and its controlled release could be extensively applied for sustainable agriculture.47 Chitosan nanocomposite and chitosan stabilized nanoparticles have been employed to develop biosensors for the detection of enterotoxigenic Escherichia coli as well as invasive Aspergillosis.48,49 However, the solubility of chitin creates a hurdle for its widespread application as it is only acid-soluble. The solubility of chitin can be enhanced by its deacetylation, creating a more soluble derivative of chitosan. Although chitosan is sparingly soluble in an aqueous medium, its chemical modifications have led to the derivation of a completely water-soluble derivative.50 TMC is one such derivative which is completely water-soluble, which makes it easier for immunizing the animal models and raising antibody response. Chitosan itself is not highly immunogenic. In our study, TMC with a high degree of quaternization, sufficiently positively charged, and the CpG ODN, a known Th1 adjuvant, was used as an adjuvant to generate a sufficient humoral response against TMC.31 CpG has been reported to be a strong Th1 adjuvant, which could even modulate an already existing Th2 response to a Th1 response.51 Hence, CpG was utilized in the study to produce a robust humoral response ensuring the production of a pronounced antibody repertoire. The negative charge present on the CpG might have aided the better electrostatic interaction with the positively charged TMC and their docketed interaction with the immune cells. The polyclonal sera against TMC were able to detect the chitin present in the fungal cell wall. These plant pathogenic fungi were detected and a fair affinity was observed between the spores and the polyclonal antibody. The chemical modification of chitin to TMC does not disrupt the rudimentary polymer backbone structure; therefore, the antibodies raised against TMC were able to detect the chitin present in fungal cell walls.
Langmuir adsorption curve revealed that the binding affinity of the polyclonal antisera was specific for chitin and the affinity could be quantitated in term of the binding coefficient as well as adsorption efficiency. The ITC-based affinity determination also illustrated that the binding was of strong quality. ITC is a sensitive technique and the affinity of binding determined is very precise. To the best of our knowledge, there is no such ITC-based study been reported.
The immunofluorescence microscopy also confirms detection of chitin all over the surface of spores, the antibodies were able to detect the surface as the chitin is present uniformly all over the cell wall. Additionally, the fungal ascospores contain up to 4.5-fold more chitosan than mycelia,52 therefore the spores were chosen for the immunofluorescence experiment. The spore is also the infective stage for fungal infection and if it could be detected, the infection can be detected with precision.53 The brightest fluorescence was detected in the Botrytis cinerea followed by Neurospora crassa and Trichoderma reesei. This could be credited to the fact that the different fungi contain different amounts of chitin in their cell wall.54–56 The chitin amount is also dependent on the variables such as strain properties, cultivation technique, and the extraction method.57 The amount of chitin present in the cell wall of Botrytis cinerea, Neurospora crassa and Trichoderma reesei has been reported by various groups as 4.8%, 4.5%, and 4.1% of the dry cell weight, respectively.58–60 These reports further support our results for the pattern of fluorescence and ELISA-based detection of chitin in the cell wall. The gamut of antibody raised against TMC must also be containing a fraction of antibody against N-acetyl glucosamine, which is the monomeric unit of the chitin. Also, the cell wall of Saccharomyces cerevisiae is made up nearly 1% glucosamine.61 This explains the trace fluorescence that was detected in Saccharomyces cerevisiae. However, this was quite faded in comparison to the other fungi used in the study.
Conclusion
The technique of generating this antibody repertoire against chitosan utilizing its methylated derivative nanoparticles is a novel attempt in the direction of development of a detection tool for fungal contamination. The downstream translation of this technology could be applied for many conditions. This polyclonal antibody has the ability to be advanced into a quality control assay, an infection detection assay, or a tool to analyze the structure of fungal cell walls. The technique could also be utilized for the detection of human fungal pathogens involved in the different fungal infections. For example, there are a number of fungi that could cause the onychomycosis, which is a nail fungal infection in human.62–66 A similar approach could be explored for other topical or dermal infections as well. The antibodies could also be conjugated with fungal inhibitors to further promote their application. In the future, the ability to target specific fungi could lead to applications in crop protection, and the containment of fungal disease outbreaks.
Acknowledgments
We would like to thank all the members of the laboratory for thoughtful and lively discussions during this work. The author HJ would like to thank the Department of Biotechnology (DBT), India. RB acknowledges DBT for funds BSLIII facility.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Latgé J-P, Beauvais A. Functional duality of the cell wall. Curr Opin Microbiol. 2014;20:111–117. doi:10.1016/j.mib.2014.05.009
2. Osumi M. The ultrastructure of yeast: cell wall structure and formation. Micron. 1998;29(2–3):207–233. doi:10.1016/S0968-4328(97)00072-3
3. Gonçalves IR, Brouillet S, Soulié M-C, et al. Genome-wide analyses of chitin synthases identify horizontal gene transfers towards bacteria and allow a robust and unifying classification into fungi. BMC Evol Biol. 2016;16(1):252. doi:10.1186/s12862-016-0815-9
4. Arbelet D, Malfatti P, Simond-Côte E, et al. Disruption of the Bcchs3a chitin synthase gene in Botrytis cinerea is responsible for altered adhesion and overstimulation of host plant immunity. Mol Plant Microbe Interact. 2010;23(10):1324–1334. doi:10.1094/MPMI-02-10-0046
5. Treitschke S, Doehlemann G, Schuster M, Steinberg G .The myosin motor domain of fungal chitin synthase V is dispensable for vesicle motility but required for virulence of the maize pathogen Ustilago maydis. Plant Cell.2010;22:2476–2494. doi:10.1105/tpc.110.075028
6. Elieh-Ali-Komi D, Hamblin MR. Chitin and chitosan: production and application of versatile biomedical nanomaterials. Int J Adv Res (Indore). 2016;4(3):411.
7. Hoell IA, Vaaje-Kolstad G, Eijsink VG. Structure and function of enzymes acting on chitin and chitosan. Biotechnol Genet Eng Rev. 2010;27(1):331–366. doi:10.1080/02648725.2010.10648156
8. Roy JC, Salaün F, Giraud S, Ferri A, Chen G. Solubility of Chitin: solvents, solution behaviors and their related mechanisms. In: Solubility of Polysaccharides. InTech; 2017.
9. Jayakumar R, Nwe N, Tokura S, Tamura H. Sulfated chitin and chitosan as novel biomaterials. Int J Biol Macromol. 2007;40(3):175–181. doi:10.1016/j.ijbiomac.2006.06.021
10. Tsigos I, Martinou A, Kafetzopoulos D, Bouriotis V. Chitin deacetylases: new, versatile tools in biotechnology. Trends Biotechnol. 2000;18(7):305–312. doi:10.1016/S0167-7799(00)01462-1
11. Kuchitsu K, Kosaka H, Shiga T, Shibuya N. EPR evidence for generation of hydroxyl radical triggered byN-acetylchitooligosaccharide elicitor and a protein phosphatase inhibitor in suspension-cultured rice cells. Protoplasma. 1995;188(1–2):138–142. doi:10.1007/BF01276805
12. Minami E, Kuchitsu K, He D-Y, et al. Two novel genes rapidly and transiently activated in suspension-cultured rice cells by treatment with N-acetylchitoheptaose, a biotic elicitor for phytoalexin production. Plant Cell Physiol. 1996;37(4):563–567. doi:10.1093/oxfordjournals.pcp.a028981
13. Libault M, Wan J, Czechowski T, Udvardi M, Stacey G. Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol Plant Microbe Interact. 2007;20(8):900–911. doi:10.1094/MPMI-20-8-0900
14. Hadwiger LA, Beckman JM. Chitosan as a component of pea-Fusarium solani interactions. Plant Physiol. 1980;66(2):205–211. doi:10.1104/pp.66.2.205
15. Köhle H, Jeblick W, Poten F, Blaschek W, Kauss H. Chitosan-elicited callose synthesis in soybean cells as a Ca2+-dependent process. Plant Physiol. 1985;77(3):544–551. doi:10.1104/pp.77.3.544
16. Malerba M, Cerana R. Chitosan effects on plant systems. Int J Mol Sci. 2016;17(7):996. doi:10.3390/ijms17070996
17. Li P, Linhardt RJ, Cao Z. Structural characterization of oligochitosan elicitor from Fusarium sambucinum and its elicitation of defensive responses in Zanthoxylum bungeanum. Int J Mol Sci. 2016;17(12):2076. doi:10.3390/ijms17122076
18. Walker-Simmons M, Ryan CA. Proteinase inhibitor synthesis in tomato leaves: induction by chitosan oligomers and chemically modified chitosan and chitin. Plant Physiol. 1984;76(3):787–790. doi:10.1104/pp.76.3.787
19. Akiyama K, Kawazu K, Kobayashi A. Partially N-deacetylated chitin oligomers (pentamer to heptamer) are potential elicitors for (+)-pisatin induction in pea epicotyls. Z Naturforsch C. 1995;50(5–6):391–397. doi:10.1515/znc-1995-5-609
20. Cord-Landwehr S, Melcher RL, Kolkenbrock S, Moerschbacher BM. A chitin deacetylase from the endophytic fungus Pestalotiopsis sp. efficiently inactivates the elicitor activity of chitin oligomers in rice cells. Sci Rep. 2016;6:38018. doi:10.1038/srep38018
21. Sahai A, Manocha M. Chitinases of fungi and plants: their involvement in morphogenesis and host‐parasite interaction. FEMS Microbiol Rev. 1993;11(4):317–338. doi:10.1111/fmr.1993.11.issue-4
22. Hamid R, Khan MA, Ahmad M, et al. Chitinases: an update. J Pharm Bioallied Sci. 2013;5(1):21. doi:10.4103/0975-7406.106559
23. Zhao Y, Park R-D, Muzzarelli RA. Chitin deacetylases: properties and applications. Mar Drugs. 2010;8(1):24–46. doi:10.3390/md8010024
24. Ride J, Barber M. Purification and characterization of multiple forms of endochitinase from wheat leaves. Plant Sci. 1990;71(2):185–197. doi:10.1016/0168-9452(90)90008-C
25. Schubert M, Agdour S, Fischer R, Olbrich Y, Schinkel H, Schillberg S. A monoclonal antibody that specifically binds chitosan in vitro and in situ on fungal cell walls. J Microbiol Biotechnol. 2010;20(8):1179–1184. doi:10.4014/jmb.1001.02023
26. Sorlier P, Hartmann DJ, Denuzière A, Viton C, Domard A. Preparation and development of anti‐chitosan antibodies. J Biomed Mater Res A. 2003;67(3):766–774. doi:10.1002/jbm.a.10132
27. Nampally M, Moerschbacher BM, Kolkenbrock S. Fusion of a novel genetically engineered chitosan affinity protein and green fluorescent protein for specific detection of chitosan in vitro and in situ. Appl Environ Microbiol. 2012;78(9):3114. doi:10.1128/AEM.07506-11
28. Li H-P, Zhang J-B, Shi R-P, Huang T, Fischer R, Liao Y-C. Engineering fusarium head blight resistance in wheat by expression of a fusion protein containing a fusarium-specific antibody and an antifungal peptide. Mol Plant Microbe Interact. 2008;21(9):1242–1248. doi:10.1094/MPMI-21-9-1242
29. Peschen D, Li H-P, Fischer R, Kreuzaler F, Liao Y-C. Fusion proteins comprising a Fusarium-specific antibody linked to antifungal peptides protect plants against a fungal pathogen. Nat Biotechnol. 2004;22:732. doi:10.1038/nbt970
30. Malik A, Gupta M, Gupta V, Gogoi H, Bhatnagar R. Novel application of trimethyl chitosan as an adjuvant in vaccine delivery. Int J Nanomedicine. 2018;13:7959. doi:10.2147/IJN.S165876
31. Malik A, Gupta M, Mani R, Gogoi H, Bhatnagar R. Trimethyl chitosan nanoparticles encapsulated protective antigen protects the mice against anthrax. Front Immunol. 2018;9:562. doi:10.3389/fimmu.2018.00562
32. Slütter B, Plapied L, Fievez V, et al. Mechanistic study of the adjuvant effect of biodegradable nanoparticles in mucosal vaccination. J Control Release. 2009;138(2):113–121. doi:10.1016/j.jconrel.2009.05.011
33. Muzzarelli R, Tarsi R, Filippini O, Giovanetti E, Biagini G, Varaldo P. Antimicrobial properties of N-carboxybutyl chitosan. Antimicrob Agents Chemother. 1990;34(10):2019–2023. doi:10.1128/AAC.34.10.2019
34. Rhoades J, Roller S. Antimicrobial actions of degraded and native chitosan against spoilage organisms in laboratory media and foods. Appl Environ Microbiol. 2000;66(1):80–86. doi:10.1128/AEM.66.1.80-86.2000
35. Jeon Y-J, Park P-J, Kim S-K. Antimicrobial effect of chitooligosaccharides produced by bioreactor. Carbohydr Polym. 2001;44(1):71–76. doi:10.1016/S0144-8617(00)00200-9
36. No HK, Park NY, Lee SH, Meyers SP. Antibacterial activity of chitosans and chitosan oligomers with different molecular weights. Int J Food Microbiol. 2002;74(1–2):65–72. doi:10.1016/S0168-1605(01)00717-6
37. Raafat D, Sahl HG. Chitosan and its antimicrobial potential–a critical literature survey. Microb Biotechnol. 2009;2(2):186–201. doi:10.1111/j.1751-7915.2008.00080.x
38. Song R-F, Li X-J, Cheng X-L, et al. Paclitaxel-loaded trimethyl chitosan-based polymeric nanoparticle for the effective treatment of gastroenteric tumors. Oncol Rep. 2014;32(4):1481–1488. doi:10.3892/or.2014.3344
39. Amidi M, Romeijn SG, Borchard G, Junginger HE, Hennink WE, Jiskoot W. Preparation and characterization of protein-loaded N-trimethyl chitosan nanoparticles as nasal delivery system. J Control Release. 2006;111(1–2):107–116. doi:10.1016/j.jconrel.2005.11.014
40. Muzzarelli RA, Tanfani F. The N-permethylation of chitosan and the preparation of N-trimethyl chitosan iodide. Carbohydr Polym. 1985;5(4):297–307. doi:10.1016/0144-8617(85)90037-2
41. Ponec V. Physical chemistry of surfaces, fifth edition. A. W. Adamson. John Wiley & Sons, Chichester, 1990. IX + 777 pp., £47.50. ISBN 0-471-61019-4. Recueil Des Travaux Chimiques des Pays-Bas. 1991;110(4):137. doi:10.1002/recl.19911100413
42. Lenardon MD, Munro CA, Gow NA. Chitin synthesis and fungal pathogenesis. Curr Opin Microbiol. 2010;13(4):416–423. doi:10.1016/j.mib.2010.05.002
43. Pisa D, Alonso R, Rábano A, Horst MN, Carrasco L. Fungal enolase, β-tubulin, and chitin are detected in brain tissue from Alzheimer’s disease patients. Front Microbiol. 2016;7:1772. doi:10.3389/fmicb.2016.01772
44. Sendid B, Dotan N, Nseir S, et al. Antibodies against Glucan, Chitin, and Saccharomyces cerevisiae Mannan as new biomarkers of Candida albicans infection that complement tests based on C. albicans Mannan. Clin Vaccine Immunol. 2008;15(12):1868.
45. Monzavi-Karbassi B, Cunto-Amesty G, Luo P, Kieber-Emmons T. Peptide mimotopes as surrogate antigens of carbohydrates in vaccine discovery. Trends Biotechnol. 2002;20(5):207–214. doi:10.1016/S0167-7799(02)01940-6
46. Baranwal A, Kumar A, Priyadharshini A, et al. Chitosan: an undisputed bio-fabrication material for tissue engineering and bio-sensing applications. Int J Biol Macromol. 2018;110:110–123. doi:10.1016/j.ijbiomac.2018.01.006
47. Kashyap PL, Xiang X, Heiden P. Chitosan nanoparticle based delivery systems for sustainable agriculture. Int J Biol Macromol. 2015;77:36–51. doi:10.1016/j.ijbiomac.2015.02.039
48. Bansal S, Jyoti A, Mahato K, Chandra P, Prakash R. Highly sensitive in vitro biosensor for enterotoxigenic escherichia coli detection based on ssDNA anchored on PtNPs‐chitosan nanocomposite. Electroanalysis. 2017;29(11):2665–2671. doi:10.1002/elan.201600169
49. Bhatnagar I, Mahato K, Ealla KKR, Asthana A, Chandra P. Chitosan stabilized gold nanoparticle mediated self-assembled glip nanobiosensor for diagnosis of invasive aspergillosis. Int J Biol Macromol. 2018;110:449–456. doi:10.1016/j.ijbiomac.2017.12.084
50. Kumirska J, Weinhold MX, Thöming J, Stepnowski P. Biomedical activity of chitin/chitosan based materials—influence of physicochemical properties apart from molecular weight and degree of N-acetylation. Polymers. 2011;3(4):1875–1901. doi:10.3390/polym3041875
51. Weeratna RD, Brazolot Millan CL, McCluskie MJ, Davis HL. CpG ODN can re-direct the Th bias of established Th2 immune responses in adult and young mice. FEMS Immunol Med Microbiol. 2001;32(1):65–71. doi:10.1111/fim.2001.32.issue-1
52. Briza P, Ellinger A, Winkler G, Breitenbach M. Chemical composition of the yeast ascospore wall. The second outer layer consists of chitosan. J Biol Chem. 1988;263(23):11569–11574.
53. Sexton AC, Howlett BJ. Parallels in fungal pathogenesis on plant and animal hosts. Eukaryot Cell. 2006;5(12):1941–1949. doi:10.1128/EC.00277-06
54. Bartnicki-Garcia S, Nickerson WJ. Isolation, composition, and structure of cell walls of filamentous and yeast-like forms of Mucor rouxii. Biochim Biophys Acta. 1962;58(1):102–119. doi:10.1016/0006-3002(62)90822-3
55. Pochanavanich P, Suntornsuk W. Fungal chitosan production and its characterization. Lett Appl Microbiol. 2002;35(1):17–21. doi:10.1046/j.1472-765X.2002.01118.x
56. White SA, Farina PR, Fulton I. Production and isolation of chitosan from Mucor rouxii. Appl Environ Microbiol. 1979;38(2):323–328.
57. Kasaai MR. Various methods for determination of the degree of N-acetylation of chitin and chitosan: a review. J Agric Food Chem. 2009;57(5):1667–1676. doi:10.1021/jf803001m
58. Soulié M-C, Piffeteau A, Choquer M, Boccara M, Vidal-Cros A. Disruption of Botrytis cinerea class I chitin synthase gene Bcchs1 results in cell wall weakening and reduced virulence. Fungal Genet Biol. 2003;40(1):38–46. doi:10.1016/S1087-1845(03)00065-3
59. Subramanyam C, Venkateswerlu G, Rao S. Cell wall composition of Neurospora crassa under conditions of copper toxicity. Appl Environ Microbiol. 1983;46(3):585–590.
60. Wang M, Zhao Q, Yang J, et al. A mitogen-activated protein kinase Tmk3 participates in high osmolarity resistance, cell wall integrity maintenance and cellulase production regulation in Trichoderma reesei. PLoS One. 2013;8(8):e72189. doi:10.1371/journal.pone.0072189
61. Lesage G, Bussey H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70(2):317–343. doi:10.1128/MMBR.00038-05
62. Baswan S, Kasting GB, Li SK, et al. Understanding the formidable nail barrier: a review of the nail microstructure, composition and diseases. Mycoses. 2017;60(5):284–295. doi:10.1111/myc.2017.60.issue-5
63. Baswan S. Transport of Charged and Uncharged Solutes in Hydrated Human Nail Plate. University of Cincinnati; 2014.
64. Baswan SM, Li SK, LaCount TD, Kasting GB. Size and charge dependence of ion transport in human nail plate. J Pharm Sci. 2016;105(3):1201–1208. doi:10.1016/j.xphs.2015.12.011
65. Baswan SM, Li SK, Kasting GB. Diffusion of uncharged solutes through human nail plate. Pharm Dev Technol. 2016;21(2):255–260. doi:10.3109/10837450.2014.991876
66. Baswan S, Kasting GB Characterization of ion transport across human nail plate. IntJ Cosmet Sci. 2012;34(4):360–360.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.