Back to Journals » OncoTargets and Therapy » Volume 8
Impact on growth and invasion of gastric cancer cell lines by silencing NEDD9
Authors Zhang S, Wu L, Liu Q, Chen K, Zhang X
Received 15 September 2014
Accepted for publication 24 November 2014
Published 20 January 2015 Volume 2015:8 Pages 223—231
DOI https://doi.org/10.2147/OTT.S74075
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 8
Editor who approved publication: Dr Jianmin Xu
Sisen Zhang,1 Lihua Wu,1 Qing Liu,1 Kuisheng Chen,2 Xiefu Zhang2
1Zhengzhou People’s Hospital Affiliated to Southern Medical University, 2The First Affiliated Hospital of Zhengzhou University, Zhengzhou, People’s Republic of China
Background: Gastric adenocarcinoma is a predominant disease with latent attribute, high malignancy, and poor prognosis in People’s Republic of China. Gastric cancer is the most common malignant tumor of the digestive tract. It has been suggested that abnormal expression of NEDD9 was associated with stage progression and metabolism of carcinomas. Some authors demonstrated that both messenger RNA (mRNA) and protein of NEDD9 were highly expressed in gastric cancer, and paired paracancerous atypical hyperplasia tissues were correlated with lymph node metastasis, tumor depth, and tumor-lymph node-metastasis (TNM) staging. In this study, we found that NEDD9 small interfering RNA (siRNA) can induce apoptosis and suppress proliferation, migration, and invasion of BGC823 cell lines. These findings suggested that NEDD9 siRNA might serve as a tumor suppressor by targeting NEDD9 in gastric cancer cell. It has been suggested that abnormal expression of NEDD9 was associated with carcinogenesis, and in the first part of the study, we found that NEDD9 was highly expressed in gastric cancer tissues; and it too was correlated with lymph node metastasis, tumor depth, and TNM staging. In this project, experiments were carried out to silence NEDD9 in BGC823 cell lines using NEDD9 siRNA, and the biological activity of BGC823 cells was observed after RNA interference.
Methods: The target analysis of NEDD9 siRNA was forecast using online tools. In order to determine a more efficient NEDD9 siRNA, three pairs of NEDD9 siRNA primer were designed, synthesized, and then transfected into BGC823 cells. NEDD9-2 siRNA was finally adopted by detecting the quantitative real-time polymerase chain reaction (qRT-PCR). Cells were collected for detecting mRNA by qRT-PCR or protein by western blot analysis. Cell apoptosis was detected using flow cytometry, and the transwell invasion system was used for cell migration and invasion assays. The effect of NEDD9 siRNA in silencing the target gene in BGC823 cells was then assessed. Also, the impact of NEDD9-2 siRNA on cell proliferation, apoptosis, migration, and invasion were detected in BGC823 cell lines.
Results: The relative quantity of expression of mRNA and protein showed a decrease in all cells transfected with siNEDD9-2 at different concentrations. The cell proliferation inhibition assay showed that the inhibition rate was significantly increased in all transfected cells compared with control groups. Cell apoptosis assay showed that the number of living cells were significantly reduced compared with control groups, and cell migration and invasion assay showed that siNEDD9 could inhibit BGC823 cell migration and invasion in vitro.
Conclusion: NEDD9 siRNA could inhibit expression of NEDD9 and induce apoptosis, suppress proliferation, migration, and invasion of BGC823 cells, acting as a tumor suppressor in carcinogenesis of gastric cancer. These findings suggested that NEDD9 siRNA plays an important role in the proliferation, apoptosis, and invasion of BGC823 cells.
Keywords: NEDD9 siRNA, apoptosis
Introduction
Gastric cancer is one of the most common cancers and frequent causes of cancer-related deaths in People’s Republic of China.1–3 The occurrence of gastric cancer is a multistage, progressive process, and its mechanism is not well understood. The diagnosis of patients is still immature despite the development of therapeutic options such as surgery, radiotherapy, and chemotherapy. Therefore, this study focuses on exploring the molecular mechanisms of gastric cancer development and the search for more effective treatments.
It has been suggested that NEDD9 plays an important role in cell proliferation, apoptosis, adhesion, migration, and invasion.4 The expression of NEDD9 is upregulated in many cancers, such as gastric cancer,5 melanoma, and glioblastoma,6,7 suggesting that the abnormal expression of NEDD9 is significantly related to the occurrence and development of cancers.
Kim et al6 found that NEDD9 is amplified and highly expressed in melanoma, and decreasing the expression of NEDD9 through RNA interference can weaken the metastasis ability of melanoma. Natarajan et al7 indicated that NEDD9 can promote the migration and invasion of cells in brain glioma, and decreasing the expression of NEDD9 through small interfering RNA (siRNA) can significantly inhibit the migration of malignant glioma cells. In addition, research on the mechanism of NEDD9 in invasion and migration of tumor showed that it might be correlated with the upregulation of NEDD9 to Src–FAK–Crk activation, and NEDD9 can interact with FAK, Src, and Crk.8,9
In the first part of the study, we demonstrated that NEDD9 was highly expressed in gastric cancer tissue.5 However, the functions of NEDD9 in gastric cancer are still not clear. It may play diverse roles in the carcinogenesis of gastric cancer. To identify whether siRNA could suppress expression of target gene specifically and exert distinct biological actions in gastric cancers, we investigated NEDD9 expression and its functional roles in gastric cancer BGC823 cell lines in this project.
Materials and methods
Design and synthesis of NEDD9 siRNA
To predict potential targets of NEDD9 siRNA, we searched GenBank, online siRNA software provided by Ambion, and BLAST, provided by the National Center for Biotechnology Information website. The three pairs of NEDD9 siRNA primer were sent to Ruibo Biological Technology Co. Ltd. (Changzhou, People’s Republic of China). Fluorescent tags were adopted to detect transfection efficiency of siRNA when they were synthesized. The NEDD9 and β-actin (as control) primers were synthesized by Sangon Biotechnology Co. Ltd. (Shanghai, People’s Republic of China). The upstream sequence of NEDD9 primer was GGGTAAAAAGGTGATAACCCCCGT; downstream: TGCTGATGAGGGAGGGATGTCGT. The length of amplified fragments of NEDD9 was 129 bp. The upstream sequence of β-actin primer was CTCCATCCTGGCCTCGCTGT; downstream: GCTGTCACCTTCACCGTTCC, and the length of amplified fragments of β-actin was 268 bp.The NEDD9 probe marked by a biotinylated 5′ end was designed and synthesized by Beijing Aoke Biotechnology Co., Ltd. (Beijing, People’s Republic of China). The sequence was 5′-GGGTAAAAAGGTGATAACCCCCGT-3 ′.
Human gastric cancer BGC823 cell lines (Cell Bank of Shanghai Institute of Life Science, Chinese Academy of Science, People’s Republic of China) were cultured in RPMI 1640, supplemented with 10% fetal bovine serum and penicillin/streptomycin in a 5% CO2 humidified incubator at 37°C (normal condition for cell culture). When the cells covered the culture flask (about 80%–90%), trypsin was added to make a single cell suspension. The cells from each flask were then divided into two or three flasks for subculturing.
Transfection of NEDD9 siRNA into BGC823 cells
In order to determine the most efficient NEDD9 siRNA on BGC823 cells, three pairs of NEDD9 siRNA primers were designed and synthesized. The BGC823 cells were divided into five groups: siNEDD9-1, siNEDD9-2, siNEDD9-3, blank control, and negative control. Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) detection found that NEDD9-2 siRNA (siNEDD9-2) inhibited the expression of NEDD9 significantly; therefore, it was selected as the best siRNA with which to carry out interference experiments. Then, three different concentrations (100, 200, or 300 nM) of siNEDD9-2 were transfected into BGC823 cells, according to the manufacturer’s instructions (lipofectamineTM2000; Thermo Fisher Scientific, Waltham, MA, USA). In addition, a blank control group (normal BGC823 cells) and a negative control group (nonspecific siRNA) were also cultured. Cells were collected for messenger RNA (mRNA) or protein assays at different time points after transfection. For apoptosis assay, cells were seeded in 6-well plates, and the transfected cells were suspended using tyrosine; a hemocytometer was used for cell counting.
RNA isolation and qRT-PCR
Total RNA in cell lines was isolated using RNAiso Plus RNA Kit (TaKaRa, Dalian, People’s Republic of China) according to the manufacturer’s instructions. In order to determine the effect of siNEDD9, total RNA was isolated at 24, 48, and 72 hours after cell transfection. Quantitation of total RNAs was performed by spectrophotometry (UNICO, Shanghai, People’s Republic of China). All samples had an excellent 260/280 ratio. RNAs were reverse transcribed into complementary DNAs (cDNAs) by using the AMV reverse transcriptase kit (TaKaRa) and were stored at −20°C for immediate or later use.
The products of qRT-PCR were electrophoresed in 1% agarose gel and detected using a Gel-doc2000 gel scan imaging analysis system. Relative expression of NEDD9 mRNA was determined according to the ratio of NEDD9 and β-actin density. The expression of β-actin was assayed for normalization of mRNA expression.
For the mRNA expression of NEDD9 and β-actin, a 50 μL reaction containing 5 μL of 10× PCR buffer, 1 μL of 10 mM deoxynucleotide mixture, 0.5 μL of Taq enzyme, 2 μL of NEDD9 or β-actin primer, 36.5 μL RNase-free water, 3 μL of 25 mM magnesium chloride, and 2 μL template cDNA. Reaction mixtures were incubated under the following conditions: 94°C for 5 minutes, 20 cycles at 94°C for 30 seconds, 62°C for 30 seconds, 72°C for 45 seconds, and finally, 72° for 7 minutes.
Western blot assay
Transfected cells were lysed in RIPA buffer kit (Sigma-Aldrich, St Louis, MO, USA) with protease inhibitors. Proteins were resolved in 10% SDS-PAGE and electrotransferred onto a nitrocellulose membrane, blocked with 5% milk for 1 hour on a decolorization shaker. After washing three times with Tris-buffered saline and Tween 20 (TBS-T), the membrane was incubated with 1:1,000 primary antibody, rat anti-human NEDD9 monoclonal antibody (Santa Cruz Biotechnology Inc., Dallas, TX, USA), at 4°C overnight. After washing three times with TBS-T, corresponding horseradish peroxidase-conjugated secondary antibody, 1:1,000, goat anti-rabbit immunoglobulin G (Wuhan Boster Biotechnology, Ltd., Wuhan, People’s Republic of China), was then used to incubate the membrane at room temperature for 1 hour. The membrane was then washed three times. Reagents in the ECL Advance western blot detection kit were blended according to the manufacturer’s instructions, and the blots were visualized in the developing solution. Bands were quantified using FluorChem FC3 software (ProteinSimple, San Jose, CA, USA) for band intensity, and β-actin was used as internal control.
Cell proliferation inhibition assay
When covering most of the culture flask (about 80%–90%), BGC823 cells were harvested and suspended into a single cell suspension using 0.25% trypsin. Then, a 100 μL volume of cell suspension was initially cultivated in 96-well plates at a density of 4.0×103 cells/well in a 5% CO2 humidified incubator at 37 °C. When cells covered the bottom of the wells, three different concentrations (100, 200, 300 nM) of siNEDD9 were added to the cells for 24, 48, and 72 hours. Nonadherent cells were removed by washing with phosphate-buffered saline (PBS). The remaining adherent cells were cultured with 20 μL of 5 mg/mL 3-(4,5-dimethylthiazol-2-yl)-2,5-di phenyltetrazolium bromide (MTT) (Sigma-Aldrich) under normal conditions. After 4 hours, the cell counting kit-8 analysis was performed for cell proliferation observation. The absorbance was detected at 490 nm in an enzyme microplate reader (Bio-Tech Instrument, USA). The growth curve was determined according to the formula: cellular proliferation inhibition rate (IR) =(1-bore experiment/optical density [OD] value contrast OD value).
Cell apoptosis assay
Cells were harvested at 72 hours after transfection, and they were suspended using 0.25% trypsin without ethylenediaminetetraacetic acid (EDTA). After washing with PBS two times, the cell concentration was adjusted to 1×106 using Annexin V-fluorescein isothiocyanate binding buffer. Cells were then suspended with 5 μL Annexin V for 15 minutes and 2.5 μL propidium iodide (4ABio, Beijing, People’s Republic of China) for 15 minutes in the dark at room temperature. After washing with PBS, 400 μL of binding buffer was added to the samples, and the analysis was performed using flow cytometry (BD Biosciences, San Jose, CA, USA). The excitation wavelength was 488 nm, and the emission wavelength was larger than 630 nm. Then, 10,000 cells were tested in each sample. At the same time, the BGC823 cells were stained with Annexin V and propidium iodide as a benchmark reference, respectively.
Cell invasion assay
The transwell invasion system was used for invasion or migration assay. After transfecting for 72 hours, cells were collected using 0.25% trypsin-EDTA (Thermo Fisher Scientific) and adjusted to 1 ×105 using RPMI 1640 without serum. The polycarbonate membrane with Matrigel matrix was plated on the upper compartment of the transwell chamber. For migration assay, the cells were added into the upper compartment of the chamber. For the invasion assay, 1×105 cells were plated on chambers precoated with Matrigel. In both assays, all cells were suspended in 100 μL RPMI 1640 without serum, and 1,000 μL of cell supernatant was added into the bottom compartment of the chamber. After 12 hours of incubation, Matrigel, dead cells, and noninvaded cells on the upper surface of the membrane were removed with a cotton swab. The membranes were fixed with 4% paraformaldehyde and stained with hematoxylin for 5 minutes. Then, the invasive cells that were stuck to the lower surface of the membrane were counted.
Statistical analysis
Statistical analysis was performed using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). All data obtained from assays were expressed as means ± standard deviation. The χ2 test was used to determine the relationship between NEDD9 expressions and control groups. Differences of quantitative data between groups were calculated. In addition, Spearman’s correlation analysis was performed to analyze the relationship between NEDD9 expression and protein expression in BGC823 cells transfected by siNEDD9. All results were considered statistically significant when P<0.05.
Results
Cell transfection
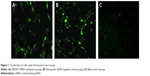
After NEDD9 siRNA and nonspecific siRNA marked by fluorescence was transfected into gastric cancer cells, BGC823, the green fluorescent protein was observed under an inverse fluorescence microscope. Significant green fluorescent protein was observed in the siRNA transfection group and the negative control group while there was no fluorescence in the blank control group. This indicated that NEDD9 had been transfected to BGC823 cells successfully. The result is shown in Figure 1.
Results of NEDD9 mRNA expression after transfection with siNEDD9-2
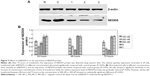
Quantitation of total RNAs were performed by spectrophotometry after total RNA was isolated. Results showed that all samples had an excellent 260/280 ratio. Then RNAs were reverse transcribed into cDNAs, and the products of qRT-PCR were electrophoresed. It showed that a band consistent with the targeted gene length in the equivalent position of 100 bp was slightly upward. Another band consistent with β-actin gene length in the equivalent location of 250 bp was slightly upward (Figure 2A).
Three different concentrations (100, 200, or 300 nM) of siNEDD9-2 were transfected into BGC823 cells. Relative expression of NEDD9 mRNA was determined according to the ratio of NEDD9 and β-actin density. The three groups of siNEDD9-2 were then compared with the control groups. Results showed that the expression of all cells decreased, except for the cells in the group transfected with 100 nM siNEDD9-2 for 24 hours (P<0.01). With the increase of concentration and extension of time of transfection, the expression level of NEDD9 mRNA was gradually reduced (P<0.01) (Figure 2B).
NEDD9 protein expression after downregulation
We analyzed expression of protein between siNEDD9-2 in different concentrations at different time points and control groups. The total proteins were extracted from groups, and the relative quantity of expression of protein was detected by western blot. Results showed that the expression of all cells transfected with siNEDD9-2 in different concentrations decreased significantly compared with control groups (P<0.05) (Figure 3A). With the increase of concentration and extension of time of transfection, the expression level of NEDD9 protein was gradually reduced (P<0.05). However, expressions between 48 hours and 72 hours in concentration of 200 nM and 300 nM showed no significant difference (Figure 3B) (P>0.05).
NEDD9-2 siRNA repressed cell proliferation and induced cell apoptosis in BGC823 cells
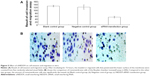
To further characterize the role of siNEDD9-2 in BGC823 cells, we observed the effects of siNEDD9-2 on proliferation and apoptosis of BGC823 cells. We compared groups 100, 200, or 300 nM for 24, 48, or 72 hours with control groups (blank or negative). Results showed that the IR increased significantly in all transfected cells. Further, the IR increased significantly with the increase of concentration and extension of transfection time (P<0.01), and the maximum IR value was observed in the 300 nM group transfected for 72 hours (Figure 4A).
Cell apoptosis was measured by flow cytometry at 72 hours after transfection. In order to explore the correlation between NEDD9 and cell apoptosis, we compared the 100, 200, or 300 nM groups with the control groups (blank and negative). Results found that the number of living cells reduced gradually (P<0.05). However, the viable and nonviable apoptotic cells increased significantly (P<0.05) (Figure4B and C).
Regulation function of siNEED9 in cell migration and invasion
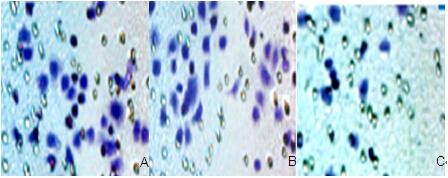
The Boyden chamber transwell assay used the BGC823 cells that were transfected with 300 nM NEDD9 siRNA for 72 hours. Compared to the control groups, the number of transmembrane cells decreased significantly, and the difference was statistically significant (P<0.001). The difference in the number of transmembrane cells between control groups had no statistical significance, as is shown in Figure 5A and B.
Discussion
Gastric cancer is one of the most common malignant tumors. The carcinogenesis and development of gastric cancer is a complex process involving multiple factors. Research on gastric cancer genes and proteins has gone to the molecular level. RNA interference technology is a new silencing technology, which involves the insertion into cells of endogenous or exogenous double stranded RNA. It can lead to the degradation of specific homologous mRNAs and the inhibition of targeted genes.
NEDD9 is a skeletal protein, belonging to the CAS (Crk-associated substrate) family. As a skeletal protein, NEDD9 acts as a router in the cell signal transduction process, although whether NEDD9 has the function of enzymology itself is unclear so far.10 Research shows that NEDD9 plays an important role in the biological characteristics of proliferation, apoptosis, adhesion, and migration of tumor cells, and those biological activities may be regulated through regulation of the expression of NEDD9 protein in tumor cells.11 Although the regulation mechanism of NEDD9 is unclear, research on melanoma and glioblastoma found that stimulating NEDD9 expression was an important event in the process of cancer metastasis.6,7
In the first part of our study, we found that the expression of NEDD9 mRNA and protein increased in turn in normal gastric mucosa tissue, atypical hyperplasia tissue adjacent to carcinoma, as well as in gastric cancer tissue, which were highly expressed at a rate of 7.5%, 20%, and 92.5%, respectively.5 The result was similar to that of a study by Kim et al.6 In addition, we identified that high expression of NEDD9 was related to in-depth tumor invasion and lymph node metastasis in gastric cancer patients, which indicated that NEDD9 may act as an oncogene and participates in the invasion and metastasis in gastric cancer.
We therefore studied expressions of NEDD9 mRNA and protein in BGC823 cells to explore the relationship between its expression and biological characteristics of gastric cancer. Results showed that mRNA and protein of NEDD9 present positive expression in BGC823 cells. The result indicated that BGC823 cell lines could be used as an in vitro research tool by which to observe the changes of cell biological characteristics after silencing NEDD9 gene.
We then designed and synthesized three pairs of siRNA targeted NEDD9 and transfected BGC823 cells by liposome to silence NEDD9 in cells. Expression of mRNA was detected by qRT-PCR and protein was detected by western blot. The results showed that the expression of NEDD9 in all cell lines transfected by siNEDD9 decreased compared with control group cells. siNEDD9-2 elicited the maximum decrease among the three cell lines; therefore, we selected siNEDD9-2 for further experiments.
We also detected cell proliferation and apoptosis using methods such as MTT and flow cytometry after silencing NEDD9 by siRNA. The results showed that both the cell growth IR and apoptosis index increased. This indicated that NEDD9 may be an important regulatory protein in cell proliferation and apoptosis of gastric cancer BGC823 cell. In addition, a study by Semizarov et al12 found that the silencing effect of siRNA to a target gene in animals could appear in 6 hours and be able to continue to 72 hours. In our study, we found that the silencing effect of siNEDD9 to the NEDD9 gene in BGC823 cells appeared at 24 hours and increased with the increase of concentration. However, the gene silencing effect showed no statistical difference at 48 and 72 hours.
Tumor metastasis is a complex process involving many factors. The process involves the change of cell polarity; the formation of microfilament, microtubules, and other important cytoskeleton protein; the regulation of kinetics of cell membrane and sticky spot; and so on. However, NEDD9 was implicated in cell adhesion and affected cell migration through the regulation of cell migration and interaction with important molecules.4 The first part of this study found that in invasive cancer of the stomach, NEDD9 presented high expression. And further, it was related to in-depth tumor invasion, lymph node metastasis, and TNM stage, suggesting that NEDD9 may play a role in metastatic gastric cancer. In this portion of our ongoing study, we transfected siNEDD9-2 into BGC823 cells and then detected invasiveness and migration capability in vitro using the Boyden invasion chamber. The results showed that the invasiveness and migration capability decreased after knockout of NEDD9. This suggests that NEDD9 plays an important role in the process of gastric cancer cell invasion.
In some studies addressing the relationship between NEDD9 and malignant tumor, Kong et al13 indicated that NEDD9 was a positive regulator of epithelial-mesenchymal transition and can promote invasion in aggressive breast cancer. Natarajan et al7 found that NEDD9 was a necessary and specific downstream effector of FAK that promotes the migration and invasion of glioblastoma cells, and Kim et al6 found that metastasis of melanoma could be inhibited by silencing NEDD9 using RNA interference technology.
Together with our clinical studies and the previous reports, these results suggest that siNEDD9 may function as a tumor suppressor by inhibiting cell proliferation, repressing migration and invasion, which in turn affected multiple clinicopathological characteristics of cancer such as lymph node metastasis and depth of invasion. Spearman correlation analysis suggested the expression of NEDD9 has a close relationship to biological characteristics of BGC823 cells. So we suggest that NEDD9 is a direct target in gastric cancer.
However, the functions of NEDD9 in gastric cancer progression remains unclear. In our study, the downward expression of NEDD9 inhibited cell proliferation in BGC823 cell lines, and interestingly, the transwell experiment also showed that siNEDD9 could inhibit BGC823 cell migration and invasion in vitro.
In conclusion, NEDD9 plays an important role in the carcinogenesis of gastric cancer. Furthermore, inhibiting the expression of NEDD9 mRNA and protein by siRNA could inhibit proliferation, migration, and invasion of BGC823 cells. This shows that NEDD9 may serve as a potential marker, and it may serve as an effective gene therapy of gastric cancer.
Acknowledgment
This study was supported by People’s Republic of China’s Key Scientific Research Project of Henan Province (132102310391).
Disclosure
The authors report no conflicts of interest in this work.
References
Forman D, Burley VJ. Gastric cancer: global pattern of the disease and an overview of environmental risk factors. Best Pract Res Clin Gastroenterol. 2006;20(4):633–649. | ||
Goh KL. Changing trends in gastrointestinal disease in the Asia-Pacific region. J Dig Dis. 2007;8(4):179–185. | ||
Jemal A, Bray F, Center MM, Ferlay J, Ward E. Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. | ||
Law SF, O’Neill GM, Fashena SJ, Einarson MB, Golemis EA. The docking protein HEF1 is an apoptotic mediator at focal adhesion sites. Mol Cell Biol. 2000;20(14):5184–5195. | ||
Sisen Z, Liu Q, Chen L, et al. The expression of NEDD9 and the relationship with invasion and metastasis in gastric cancer tissue. Chin J General Surg. 2013;11:894–895. | ||
Kim M, Gans JD, Nogueira C, et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125(7):1269–1281. | ||
Natarajan M, Stewart JE, Golemis EA, et al. HEF1 is a necessary and specific downstream effector of FAK that promotes the migration of glioblastoma cells. Oncogene. 2006;25(12):1721–1732. | ||
Law SF, Eetojak J, Wang B, Mysliwiec T, Kruh G, Golemis EA. Human enhancer of filamentation 1, a nover p130cas-like docking protein, associates with focal adhesion kinase and induces pseudohyphal growth in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16(7):3327–3337. | ||
Minegishi M, Tachibana K, Sato T, Iwata S, Nojima Y, Morimoto C. Structure and function of Cas-L, a 105-kD Crk-associated substrate-related protein that is involved in beta 1 integrin-mediated signaling in lymphocytes. J Exp Med. 1996;184(4):1365–1375. | ||
Singh M, Cowell L, Seo S, O’Neill G, Golemis E. Molecular basis for HEF1/NEDD9/Cas-L action as a multifunctional co-ordinator of invasion, apoptosis and cell cycle. Cell Biochem Biophys. 2007;48(1):54–72. | ||
Singh M, Cowell L, Seo S, O’Neill G, Golemis E. Molecular basis for HEF1/NEDD9/Cas-L action as a multifunctional co-ordinator of invasion, apoptosis and cell cycle. Cell Biochem Biophys. 2007;48(1):54–72. | ||
Semizarov D, Frost L, Sarthy A, Kroeger P, Halbert DN, Fesik SW.Specificity of short interfering RNA determined through gene expression signatures. Proc Natl Acad Sci U S A. 2003;100(11):6347–6352. | ||
Kong C, Wang C, Wang L, et al. NEDD9 is a positive regulator of epithelial-mesenchymal transition and promotes invasion in aggressive breast cancer. PLoS One. 2011;6(7):e22666. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.