Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 14
Immunogenetic Prediction of VDR Gene SNPs: Lack of Association with Susceptibility to Type 1 Diabetes in Jordanian Patients
Authors Khdair SI , Jarrar YB , Jarrar W
Received 8 August 2021
Accepted for publication 9 September 2021
Published 21 September 2021 Volume 2021:14 Pages 4079—4087
DOI https://doi.org/10.2147/DMSO.S333005
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ming-Hui Zou
Sawsan I Khdair, Yazun B Jarrar, Wassan Jarrar
Department of Pharmacy, Faculty of Pharmacy, Al-Zaytoonah University of Jordan, Amman, 11733, Jordan
Correspondence: Sawsan I Khdair
Department of Pharmacy, Faculty of Pharmacy, Al-Zaytoonah University of Jordan, Amman, 11733, Jordan
Tel +962795513275
Email [email protected]
Purpose: The interaction of Vitamin D and its receptor plays a crucial role in immune modulation. Therefore, the relationship between the pathogenesis of type 1 diabetes and the genetic variants of Vitamin D receptor, which is involved in the activity of Vitamin D, was studied extensively in different populations. The association of Vitamin D receptor gene polymorphisms with predisposition to type 1 diabetes revealed controversial and inconclusive results. The aim of this study was to examine the association of four Vitamin D receptor polymorphisms with type 1 diabetes in Jordanian patients.
Patients and Methods: Analysis of the single nucleotide polymorphisms FokI (rs2228570), ApaI (rs7975232), TaqI (rs731236) and BsmI (rs1544410) in 100 Jordanian volunteers (50 control and 50 Type 1 diabetes patients) was performed using the highly specific New Generation Sequencing technology.
Results: The distribution of allele, genotype as well as haplotype frequencies exhibited no significant (P > 0.05) differences between type 1 diabetes patients and controls. Furthermore, no differences (P > 0.05) in the frequency of the genotypes of the Vitamin D receptor genetic variants were found in relation to the age of disease onset.
Conclusion: These findings suggest these four single nucleotide polymorphisms of the Vitamin D receptor gene seem not to be associated with type 1 diabetes predisposition in Jordanian patients. Further wide genome studies are recommended to detect other genetic variant associations with type 1 diabetes among Jordanians.
Keywords: jordan, diabetes, single nucleotide polymorphism, vitamin D receptor, genetic predisposition
Introduction
Type 1 diabetes (T1D) is considered one of the worldwide prevalent autoimmune diseases, which is characterized by the destruction of insulin-producing beta cells of the pancreas.1 The prevalence of diabetes is rising globally as more than 1.1 million young people under the age of 20 live with T1D to date.2 According to the estimations of the International Diabetes Federation (IDF), 149,400 individuals under the age of 19 years had T1D in the Middle East and North African Region (MENA) in 2019.2 Jordan is one of the MENA countries but unfortunately there are no accurate numbers describing the incidence and prevalence of T1D among Jordanians due to the lack of T1D epidemiologic studies in Jordan. T1D is a multifactorial chronic disease in which the interaction of genetic, environmental, and epigenetic factors plays a crucial role. Environmental factors, nutrition, viral infections, and lifestyle habits could explain, at least in part, the increase in T1D prevalence worldwide, specifically in individuals with genetic predisposition to the disease.3–5 Several genes are linked to the development of T1D such as Human Leukocyte Antigen (HLA) genes, which are considered the strongest genetic risk factors for T1D.6 However, among the non-HLA risk genes reported so far, the Vitamin D Receptor (VDR) gene was investigated extensively as a candidate risk gene for T1D.7 The VDR is located on chromosome 12 and is comprised of eight protein-encoding exons (exons 2–9) and six untranslated exons (exons 1a-1f).8,9 VDR is a nuclear steroid receptor which binds to vitamin D and regulates the transcription of several genes related to inflammation and immunomodulation. VDR regulates the expression of target genes by specifically binding to their promoter region.10 VD acts in the immune system by suppressing the production of some cytokines such as tumor necrosis factor (TNF), interleukin (IL)-2, IL-12, and interferon. On the other hand, VD is also able to activate the expression of other cytokines such as IL-4 and transforming growth factor-1 which in turn leads to the activation of regulatory T-cells and the downregulation of T-helper 1 cells.11 VD was also shown to play a role in adaptive immunity by reducing the expression of Major Histocompatibility Complex (MHC) - class II molecules on antigen-presenting cells thus regulating T-cell–mediated immune responses.3 VDR has been found in more than thirty distinct tissues such as macrophages, dendritic cells as well as pancreatic cells.11 The activity of this receptor can be modulated by the presence of Single Nucleotide Polymorphisms (SNPs) in the VDR.12 Thus, these SNPs are suggested to play a crucial role in the activity of vitamin D that may affect the risk of T1D development.3 The most reported SNPs of the VDR are: FokI (rs2228570), ApaI (rs7975232), TaqI (rs731236) and BsmI (rs1544410).13 The importance of these polymorphic genetic variants has been investigated in association with T1D in several populations. Several studies showed an association between some of these SNPs and an increased risk of T1D development,14–19 while other studies showed no association between these SNPs and T1D.18,20–23 Therefore, the VDR polymorphisms that predispose to the development of T1D are still controversial. In view of these inconsistent results in different populations, further studies are needed to clarify the relation between VDR genetic variants and T1D development. Accordingly, the aim of this study was to investigate the association of VDR polymorphisms with T1D in Jordanian patients. Identifying the VDR genetic variants of T1D Jordanian patients may help in future in understanding the role of VDR polymorphisms in the pathogenesis of T1D.
Materials and Methods
Sample Recruitment
One hundred unrelated volunteers took part in this study; fifty healthy controls as well as fifty individuals which were diagnosed with T1D were recruited at the endocrinology department of King Abdullah University Hospital. All T1D patients were diagnosed according to the criteria of American Diabetes Associations (ADA) guidelines for Type 1 diabetes, such as testing the islets autoantibodies. T1D patients are undergoing insulin treatment, and a personalized medical nutrition therapy is also provided, exercises are also recommended. Being a Jordanian was set as an inclusion criterion for all study participants, as participants who were not of Jordanian origin were excluded from this study. Volunteers of the control group with first-degree relatives which have T1D were excluded from this study. An exclusion criterion for the T1D group was that T1D patients with other chronic diseases such as cardiovascular diseases, liver disease, and autoimmune diseases other than T1D were not included in this study. The mean age and male/female ratio of control participants were 24.7 years and 1.63, respectively. Whereas the mean age and male/female ratio of diabetes patients were 20 years and 0.47, respectively. However, the mean age of T1D onset was 10.3 years. The clinical parameters of the volunteers are listed in Table 1. Blood samples were collected from participants after signing a consent form (IRB was obtained from the ethical committee of King Abdullah University Hospital; approval number 46/117/2018). This study complied with the Declaration of Helsinki.24
 |
Table 1 Clinical Parameters of the Volunteers |
DNA Extraction and VDR Genotyping
Extraction of DNA was performed using the Wizard® Genomic DNA purification kit (Promega, USA). VDR genotyping was performed using amplicon sequencing with the Miseq Illumina platform. In this method, oligonucleotides are designed to amplify the VDR regions of the SNPs FokI, (rs2228570), ApaI (rs7975232), TaqI (rs731236) and BsmI (rs1544410) (Table 2), followed by next-generation sequencing (NGS) performed by Genochem World SL. (Spain). Quality of PCR products was assessed by capillary electrophoresis in the QIAxcel Advanced System (Qiagen). The MiSeq Reagent Kit v2 (300-cycle) on Miseq (Illumina platform) was used for sequencing. The four SNPs analyzed in our study were named according to the restriction endonucleases which were used originally for their identification using the Restriction Fragment Length Polymorphism (RFLP) method.25 The FokI T>C genetic variant (rs2228570) is located in exon 2, the BsmI A>G genetic variant (rs1544410) and the ApaI T>G genetic variant (rs7975232) are located in intron 8, and the TaqI T>C genetic variant (rs731236) in Exon 9.26
 |
Table 2 Primers Used for Amplifying Specific VDR Regions of Interest. Primer Name, Sequence, Size of PCR Fragment |
Linkage Disequilibrium and Haplotype Distribution
Linkage disequilibrium (LD) and haplotype analysis, of VDR genetic variants, for control volunteers and T1D patients was performed using the Haploview (v. 4.2) software. Measurement of LD between the VDR variants was done by Dʹ and correlation coefficient (r2).
Statistical Analysis
The Statistical Package for Social Sciences (SPSS) software was used to conduct the Statistical analysis (IBM analytics, USA). Comparison of allele, genotype, and haplotype frequencies between T1D patients and control volunteers was performed using the Chi-square (χ2) test. Deviation from Hardy-Weinberg equation, including comparison between the observed and expected number of VDR genotypes, was tested using χ2. Evaluating the association between VDR genetic variants and the sex as well as age of disease onset of volunteers was done using Chi-square (χ2) test. Binary logistic regression analysis was used to calculate the odds ratios (ORs) and 95% confidence intervals (CIs) to investigate the possible association VDR SNPs with T1D. When the p-value was less than 0.05, the results were considered significant.
Results
Our findings show allele (Table 3), genotype (Table 4) and haplotype (Table 5) frequencies of the VDR SNPs. All genotype frequencies of the VDR were in Hardy–Weinberg equilibrium as shown in Supplementary Table 1. Our results showed that there is no statistical difference in allele and genotype frequencies between T1D patients and controls (Tables 3 and 4). In addition, the co-dominant, dominant and recessive genotyping models showed no statistical difference among all studied groups (Table 4). Comparison of VDR haplotype frequencies also showed no significant (P > 0.05) differences between T1D and control volunteers (Table 5). The VDR haplotype FokI (C)/ TaqI (T)/ BsmI (G)/ ApaI (G) was slightly higher among the control in comparison to T1D group. This difference, however, was not statistically significant (P=0.06) (Table 5). In addition, although there is an obvious difference in the ratio of male/female among T1D patients and controls, our findings showed no proof of a genotypic association (P > 0.05) of all four SNPs of the VDR with T1D in relation to sex (Table 6). Furthermore, the age of disease onset did not influence (P > 0.05) the susceptibility to T1D in Jordanian patients (Table 7). Regarding the LD of VDR variants (Figure 1), this study revealed that ApaI is in a complete LD (D’=1) with BsmI and TaqI in both control and T1D volunteers. The TaqI variant was in a strong LD among both control (D’= 0.86) and T1D (D’=0.91) volunteers. However, no difference in LD of VDR variants between the tested groups was found.
 |
Table 3 VDR Allele Frequencies of T1D Patients and Controls |
 |
Table 4 The VDR Genotype Among T1D Patients and Controls |
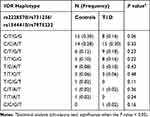 |
Table 5 The VDR Haplotype Among Type 1 Diabetic Patients and Controls |
 |
Table 6 The Association of Patients’ VDR Genotype with T1D in Relation with Sex |
 |
Table 7 The Association of VDR Genotype with the Age of T1D Onset |
Discussion
Recent advances in molecular profiling using sequencing are considered an efficient tool for developing targeted therapy for autoimmune diseases as well as a strategic tool for early diagnosis and prevention.27
VDR polymorphisms have been analyzed for their association with autoimmune diseases in different populations.28–31 However, association of VDR genetic variants with T1D were inconsistent in various ethnic groups. We conducted this study on Jordanian T1D patients to clarify the role of the well-characterized VDR genetic variants in genetic susceptibility to T1D, as no such studies were done so far in Jordan. Genotyping was done using NGS, which is known to be a highly accurate sequencing technique.32 Our results show no statistical differences among all the four well-known SNPs of the VDR: FokI T>C (rs2228570), BsmI A>G (rs1544410), ApaI T>G (rs7975232), and TaqI T>C (rs731236) among T1D patients and controls. However, our findings are in line with other studies which also showed no association between the different SNPs of the VDR and T1D in different populations such as the Portuguese,33 Chile,22 Iranian,18 Turkish,20 Romanian,34 Finnish,21 British, Norwegian and US population.35 On the other hand, populations from the MENA region such as Saudi Arabia showed a difference in the frequency of one of the VDR SNPs, namely the frequency of BsmI in T1D patients compared to controls.36 In addition, the Kuwaiti population detected significant differences in the frequency of two SNPs of the VDR, namely FokI and TaqI.17 Furthermore, associations of most or all of the four SNPs with T1D were reported in some populations such as the Pakistani,16 Greek and Japanese populations.15,19 In addition, although our results revealed no difference in LD of VDR variants between the two groups analyzed, our results showed that ApaI is in a complete LD (D’=1) with BsmI and TaqI in both control and T1D volunteers, which is in line with previous studies which showed that ApaI, BsmI, and TaqI polymorphisms are in strong LD at the 3ʹend of the VDR.37,38
Although associations of VDR polymorphisms with T1D were extensively studied in different populations, the role of these SNPs in T1D development is still unclear. These conflicting findings from various populations might be explained by ethnic differences and geographic variations among different populations which could affect the frequencies of VDR polymorphisms.25,39,40 In addition, genetic susceptibility alone is not enough to induce T1D as the interaction of different other factors plays an essential role such as environmental factors, nutrition, viral infections, and lifestyle habits.4,5
Concerning the power of statistical analysis, we must acknowledge that the sample size (n=50 for control group and n=50 for T1D group), with a total of 100 used is limited. However, the obviously robust effect of our results regarding the allele, genotype and haplotype frequencies give confidence in the main conclusion presented here. The results of our study provide an additional insight for a controversial issue, namely the association of VDR polymorphisms with predisposition to T1D. However, the implication of VDR genetic variants with T1D is still questionable in a lot of populations, so more research in the future with larger sample sizes and larger genetic scale trials are needed.
Conclusions
In conclusion, our findings revealed that VDR genetic variants have no impact on the predisposition to T1D among Jordanian patients. Further genomic studies are recommended to detect other genetic polymorphism associations with T1D among Jordanians. This type of study highlights the necessity of global genome screening to increase the number of case-control studies worldwide thereby enriching immunotherapy for precision medicine by increasing data about different genetic variants and their association to autoimmune diseases. Moreover, previous studies showed that there is a link between glycemic control of diabetic patients and other genetic variants.41 Therefore, future wide genome studies are recommended to find out novel SNPs associated with glycemic control in diabetic patients, which could increase our understanding of the pathogenesis of diabetes and help in providing personalized therapy for patients.
Abbreviations
T1D, Type 1 diabetes; IDF, International Diabetes Federation; MENA, Middle East and North African Region; HLA, Human Leukocyte Antigen; VDR, Vitamin D Receptor; TNF, tumor necrosis factor; IL, interleukin; MHC, Major Histocompatibility Complex; SNPs, Single Nucleotide Polymorphisms; ADA, American Diabetes Associations; IRB, Institutional Review Board; NGS, next-generation sequencing; RFLP, Restriction Fragment Length Polymorphism; LD, Linkage disequilibrium; SPSS, Statistical Package for Social Sciences.
Data Sharing Statement
The data used in this study are available from the corresponding author on reasonable request.
Acknowledgments
We wish to thank Genochem World SL. (Spain) for their technical support. We also thank all the volunteers who took part in this study and Ms. Safa’a Bataineh for her help in collecting blood samples. We would also like to thank Dr. Alaa Hammad for her support during the revision process.
Funding
This study was supported by a grant from Al-Zaytoonah University of Jordan [Grant number: 31/11/2020-2021].
Disclosure
The authors report no conflicts of interest in this work.
References
1. Edner NM, Heuts F, Thomas N, et al. Follicular helper T cell profiles predict response to costimulation blockade in type 1 diabetes. Nat Immunol. 2020;21(10):1244–1255. doi:10.1038/s41590-020-0744-z
2. International Diabetes Federation. IDF Diabetes Atlas; 2019. Available from: https://diabetesatlas.org/upload/resources/material/20200302_133351_IDFATLAS9e-final-web.pdf.
3. Infante M, Ricordi C, Sanchez J, et al. Influence of vitamin D on islet autoimmunity and beta-cell function in type 1 diabetes. Nutrients. 2019;11(9):1–30. doi:10.3390/nu11092185
4. Dotta F, Censini S, Van Halteren AGS, et al. Coxsackie B4 virus infection of β cells and natural killer cell insulitis in recent-onset type 1 diabetic patients. Proc Natl Acad Sci U S A. 2007;104(12):5115–5120. doi:10.1073/pnas.0700442104
5. Knip M, Simell O. Environmental triggers of type 1 diabetes. Cold Spring Harb Perspect Med. 2012;2:a007690. doi:10.1101/cshperspect.a007690
6. Todd JA. Etiology of type 1 diabetes. Immunity. 2010;32(4):457–467. doi:10.1016/j.immuni.2010.04.001
7. Zhai N, Bidares R, Makoui MH, et al. Vitamin D receptor gene polymorphisms and the risk of the type 1 diabetes: a meta-regression and updated meta-analysis. BMC Endocr Disord. 2020;20(1):1–21. doi:10.1186/s12902-020-00575-8
8. Crofts LA, Hancock MS, Morrison NA, Eisman JA. Multiple promoters direct the tissue-specific expression of novel N-terminal variant human vitamin D receptor gene transcripts. Proc Natl Acad Sci U S A. 1998;95(18):10529–10534. doi:10.1073/pnas.95.18.10529
9. Taymans SE, Pack S, Pak E, et al. The human vitamin D receptor gene (VDR) is localized to region 12cen- q12 by fluorescent in situ hybridization and radiation hybrid mapping: genetic and physical VDR map. J Bone Miner Res. 1999;14(7):1163–1166. doi:10.1359/jbmr.1999.14.7.1163
10. Christakos S, Dhawan P, Verstuyf A, Verlinden L, Carmeliet G. Vitamin D: metabolism, molecular mechanism of action, and pleiotropic effects. Physiol Rev. 2015;96(1):365–408. doi:10.1152/physrev.00014.2015
11. Jones G, Strugnell SA, DeLuca HF. Current understanding of the molecular actions of vitamin D. Physiol Rev. 1998;78(4):1193–1231.
12. Abouzid M, Karazniewicz-Lada M, Glowka F. Genetic determinants of vitamin D-related disorders; focus on vitamin D receptor. Curr Drug Metab. 2018;19(12):1042–1052. doi:10.2174/1389200219666180723143552
13. Uitterlinden AG, Fang Y, Van Meurs JB, Pols HA, Van Leeuwen JP. Genetics and biology of vitamin D receptor polymorphisms. Gene. 2004;338(2):143–156. doi:10.1016/j.gene.2004.05.014
14. Wang G, Zhang Q, Xu N, et al. Associations between two polymorphisms (FokI and BsmI) of vitamin D receptor gene and type 1 diabetes mellitus in Asian population: a meta-analysis. PLoS One. 2014;9(3):e89325.
15. Panierakis C, Goulielmos G, Mamoulakis D, Petraki E, Papavasiliou E, Galanakis E. Vitamin D receptor gene polymorphisms and susceptibility to type 1 diabetes in Crete, Greece. Clin Immunol. 2009;133(2):276–281. doi:10.1016/j.clim.2009.08.004
16. Mukhtar M, Batool A, Wajid A, Qayyum I. Vitamin D receptor gene polymorphisms influence T1D susceptibility among Pakistanis. Int J Genomics. 2017;2017:1–6. doi:10.1155/2017/4171254
17. Rasoul MA, Haider MZ, Al-Mahdi M, Al-Kandari H, Dhaunsi GS. Relationship of four vitamin D receptor gene polymorphisms with type 1 diabetes mellitus susceptibility in Kuwaiti children. BMC Pediatr. 2019;19(1):1–13. doi:10.1186/s12887-019-1448-0
18. Mohammadnejad Z, Ghanbari M, Ganjali R, et al. Association between vitamin D receptor gene polymorphisms and type 1 diabetes mellitus in Iranian population. Mol Biol Rep. 2012;39(2):831–837. doi:10.1007/s11033-011-0805-3
19. Ban Y, Taniyama M, Yanagawa T, et al. Vitamin D receptor initiation codon polymorphism influences genetic susceptibility to type 1 diabetes mellitus in the Japanese population. BMC Med Genet. 2001;2. doi:10.1186/1471-2350-2-7
20. Gogas Yavuz D, Keskin L, KIyIcI S, et al. Vitamin D receptor gene BsmI, FokI, ApaI, TaqI polymorphisms and bone mineral density in a group of Turkish type 1 diabetic patients. Acta Diabetol. 2011;48(4):329–336. doi:10.1007/s00592-011-0284-y
21. Turpeinen H, Hermann R, Vaara S, et al. Vitamin D receptor polymorphisms: no association with type 1 diabetes in the Finnish population. Eur J Endocrinol. 2003;149(6):591–596. doi:10.1530/eje.0.1490591
22. Angel B, Santos JL, Carrasco E, Albala C, Pérez-Bravo F. Vitamin D receptor polymorphism and susceptibility to type 1 diabetes in Chilean subjects: a case-parent study. Eur J Epidemiol. 2004;19(12):1085–1087. doi:10.1007/s10654-004-1026-z
23. Thorsen SU, Mortensen HB, Carstensen B, et al. No association between type 1 diabetes and genetic variation in vitamin D metabolism genes: a Danish study. Pediatr Diabetes. 2014;15(6):416–421. doi:10.1111/pedi.12105
24. World Medical Association. World medical association declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191–2194. doi:10.1001/jama.2013.281053
25. Tizaoui K, Kaabachi W, Hamzaoui A, Hamzaoui K. Contribution of VDR polymorphisms to type 1 diabetes susceptibility: systematic review of case – control studies and meta-analysis. J Steroid Biochem Mol Biol. 2014;143:240–249. doi:10.1016/j.jsbmb.2014.03.011
26. Penna-Martinez M, Badenhoop K. Inherited variation in vitamin D genes and type 1 diabetes predisposition. Genes. 2017;8(4):2–9. doi:10.3390/genes8040125
27. Ma Y, Shi N, Li M, Chen F, Niu H. Applications of next-generation sequencing in systemic autoimmune diseases. Genom Proteom Bioinform. 2015;13(4):242–249. doi:10.1016/j.gpb.2015.09.004
28. Kriegel MA, Manson JE, Costenbader KH. Does vitamin D affect risk of developing autoimmune disease?: A systematic review. Semin Arthritis Rheum. 2011;40(6):512–531. doi:10.1016/j.semarthrit.2010.07.009
29. Aydngz IE, Bingül I, Doǧru-Abbasoǧlu S, Vural P, Uysal M. Analysis of vitamin D receptor gene polymorphisms in vitiligo. Dermatology. 2012;224(4):361–368. doi:10.1159/000339340
30. Lin WY, Wan L, Tsai CH, Chen RH, Lee CC, Tsai FJ. Vitamin D receptor gene polymorphisms are associated with risk of Hashimoto’s thyroiditis in Chinese patients in Taiwan. J Clin Lab Anal. 2006;20(3):109–112. doi:10.1002/jcla.20110
31. Mukhtar M, Sheikh N, Suqaina SK, et al. Vitamin D receptor gene polymorphism: an important predictor of arthritis development. Biomed Res Int. 2019;2019:1–8. doi:10.1155/2019/8326246
32. Behjati S, Tarpey PS. What is next generation sequencing? Arch Dis Child Educ Pract Ed. 2013;98(6):236–238. doi:10.1136/archdischild-2013-304340
33. Lemos MC, Fagulha A, Coutinho E, et al. Lack of association of vitamin D receptor gene polymorphisms with susceptibility to type 1 diabetes mellitus in the Portuguese population. Hum Immunol. 2008;69(2):134–138. doi:10.1016/j.humimm.2008.01.008
34. Guja C, Marshall S, Welsh K, et al. The study of CTLA-4 and vitamin D receptor polymorphisms in the Romanian type 1 diabetes population. J Cell Mol Med. 2002;6(1):75–81. doi:10.1111/j.1582-4934.2002.tb00312.x
35. Nejentsev S, Cooper JD, Godfrey L, et al. Analysis of the vitamin D receptor gene sequence variants in type 1 diabetes. Diabetes. 2004;53(10):2709–2712. doi:10.2337/diabetes.53.10.2709
36. Iyer A, Lanham-New S, Khoja S, Al-Ghamdi M, Al Doghaither H. Relationship between vitamin d receptor gene polymorphisms and type 1 diabetes mellitus in Saudi patients. Int J Pharmacol. 2017;13(8):1092–1097. doi:10.3923/ijp.2017.1092.1097
37. Zmuda JM, Cauley JA, Ferrell RE. Molecular epidemiology of vitamin D receptor gene variants. Epidemiol Rev. 2000;22(2):203–217. doi:10.1093/oxfordjournals.epirev.a018033
38. Thakkinstian A, D’Este C, Attia J. Haplotype analysis of VDR gene polymorphisms: a meta-analysis. Osteoporos Int. 2004;15(9):729–734. doi:10.1007/s00198-004-1601-x
39. Sahin OA, Goksen D, Ozpinar A, Serdar M, Onay H. Association of vitamin D receptor polymorphisms and type 1 diabetes susceptibility in children: a meta-analysis. Endocr Connect. 2017;6(3):159–171. doi:10.1530/EC-16-0110
40. Savastio S, Cadario F, Beux S, et al. Vitamin D and type I diabetes. Open Rheumatol J. 2018;12(1):289–299. doi:10.2174/1874312901812010289
41. Kassem DH, Adel A, Sayed GH, Kamal MM. A novel SERPINB1 single-nucleotide polymorphism associated with glycemic control and β-cell function in Egyptian type 2 diabetic patients. Front Endocrinol. 2020;11:1–8. doi:10.3389/fendo.2020.00450
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

