Back to Journals » Cancer Management and Research » Volume 13
Imbalance of Molecular Module of TINCR-miR-761 Promotes the Metastatic Potential of Early Triple Negative Breast Cancer and Partially Offsets the Anti-Tumor Activity of Luteolin
Authors Zhang ML, Liu WW, Li WD
Received 22 October 2020
Accepted for publication 10 December 2020
Published 23 February 2021 Volume 2021:13 Pages 1877—1886
DOI https://doi.org/10.2147/CMAR.S288271
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Man-Li Zhang, Wei-Wei Liu, Wei-Dong Li
Department of Breast Surgery, Cangzhou People’s Hospital, Cangzhou City, 061001, Hebei Province, People’s Republic of China
Correspondence: Man-Li Zhang
Department of Breast Surgery, Cangzhou People’s Hospital, Intersection of Chongqing Road and Jilin Avenue, Yunhe District, Cangzhou City, 061001, Hebei Province, People’s Republic of China
Tel +86-15832714444
Email [email protected]
Background: Triple negative breast cancer (TNBC) poses a great threat to patient prognosis. LncRNA-miRNA is a molecular module formed by a long non-coding RNA (LncRNA) and a microRNA (miRNA) that mediates the metastatic potential of tumours such as TNBC, and luteolin (LU) is a natural compound with anti-TNBC activity.
Objective: We aim to explore the regulatory mechanism of terminal differentiation-induced non-coding RNA (TINCR)-miR-761 molecular module in early TNBC, as well as its influence on anti-tumor activity of LU.
Methods: The serum was collected from TNBC patients in early stage to detect the expression of TINCR and miR-761 using RT-PCR. Transwell method was applied for the determination of cell migration and invasion, Western blot for epithelial–mesenchymal transition (EMT), flow cytometry (FCM) for cell apoptosis, and dual luciferase reporter and RNA pull-down experiment for the verification of the targeted relationship between TINCR and miR-761.
Results: Both TINCR and miR-761 were up-regulated in the serum of patients with early TNBC and the area under the curve (AUC) of the two for distinguishing TNBC from BC was not less than 0.850. In the cell function tests, down-regulation of TINCR or miR-761 notably suppressed the metastatic potentials (cell migration, invasion and EMT) of TNBC cells were remarkably inhibited, while up-regulation of TINCR or miR-761 notably promoted the metastatic potentials. We also confirmed that TINCR acts as the molecular sponge of miR-761, and has positive regulation on it. Besides, LU can significantly down-regulate TINCR and miR-761, and partially offset the anti-TNBC activity of LU when they were abnormally up-regulated, which was mainly reflected in the decrease of anti-proliferation and pro-apoptotic ability of LU against TNBC.
Conclusion: There is an imbalance of TINCR-miR-761 molecular module in early TNBC, which may be a potential new therapeutic target of TNBC.
Keywords: triple negative breast cancer, TINCR, miR-761, metastasis, luteolin
Introduction
Breast cancer (BC) is one of the most common cancers among women aged 40–45 years, and is also one of the important causes of cancer-related deaths among women.1 At present, the risk of BC is still on the rise, and triple negative breast cancer (TNBC), as a highly invasive phenotype of BC, has a higher risk of death.2,3 The main features of TNBC are high recurrence rate, high metastasis rate and high mortality rate, and its tumor metastasis potential has a key impact on the prognosis of early TNBC patients.4,5 Luteolin (LU), a natural flavonoid, has certain anti-tumor activity in various cancers. It has been proved to be able to inhibit the migration, invasion and epithelial–mesenchymal transition (EMT) of TNBC cells by down-regulating the mechanism of TNBC metastasis mediated by β-catenin.6 In this study, we first explored potential serum-based specific bioindicators in early-stage TNBC patients, and then evaluated TNBC metastatic potential, and the impact of anti-tumor activity of LU in proliferation and apoptosis based on the aforementioned indicators. Our research may help to distinguish early TNBC patients from early BC patients, and may provide new insights for the treatment of early TNBC.
The exploration of long non-coding RNA-micro RNA (LncRNA-miRNA) molecular module can help reveal the occurrence and development of various physiological or pathological processes. LncRNA is equivalent to a long-chain nucleotide module, which can play a regulatory role in tumor progression by targeting short-chain nucleotide modules such as miRNA.7–9 LncRNA-miRNA molecular module has been proved to play a certain role in the metastasis potential of TNBC. For example, the interaction between human metastasis-associated lung adenocarcinoma transcript 1 (MALAT1)-miR-1 molecular module can regulate the metastasis of TNBC cells.10 Another example is that the molecular module of growth arrest specific transcription factor 5 (GAS5)-miR-196a-5p mediates the invasive molecular phenotype and lymph node metastasis of TNBC.11 Terminal differentiation-induced non-coding RNA (TINCR) is a carcinogenic lncRNA related to BC and TNBC, whose repression knock-down can reverse the progress of BC via cell survival inhibition.12 In addition, there is a certain indirect relationship between TINCR and LU, both of them can act as epigenetic regulatory factors and epigenetic regulators in the process of epidermal differentiation.13 MiR-761 can induce invasive phenotype of TNBC. Its up-regulation facilitates cell migration, invasion and lung metastasis of TNBC.14
We found potential binding sites between TINCR and miR-761 via online target gene prediction website. Based on the above research, we suspected that the abnormality of TINCR-miR-761 molecular module may respond to early TNBC, and may have certain impacts on the metastatic potential of TNBC cells and the anti-TNBC effect of LU, which is hereby verified later.
Data and Methods
Serum Collection of TNBC and BC Patients
This study was conducted according to the principles of Declaration of Helsinki, and was approved by the Ethics Committee of Cangzhou People’s Hospital. All subjects provided informed consent and knew the purpose of the study. Serum samples were collected from 50 patients with early stage (I–II) TNBC and 40 patients with early stage (I–II) BC in Cangzhou People’s Hospital from January 2010 to December 2019. None of these subjects were complicated with other malignant tumors, severe organ dysfunction, other breast diseases or infectious diseases, and all of them were treated for the first time.
Cell Culture and Transfection
Our human normal mammary epithelial cells MCF-10A and TNBC cells MDA-MB-468, BT-549, SUM159PT, HCC1937 were commercially purchased from Otwo Biotech (Otwo Biotech Co., Ltd., Shenzhen, China, HT-X1884, HT-X1638, HT-X1642, HTX-2782, HT-X1644). Cells were placed in RPMI 1640 medium (Jingke Chemical Technology Co., Ltd., Shanghai, China, GNM-11,835) in a humid environment at 37°C, with 5%CO2 and 95% air. The medium contained 10% (v/v) fetal bovine serum (FBS) (Lianshuo Biological Technology Co., Ltd., Shanghai, China, A3160801), 100 U/mL penicillin and 100 μg/mL streptomycin (Noble Ryder Technology Co., Ltd., Beijing, China, 15,240,062). The incubation was conducted under the intervention of LU (Baoman Biotechnology Co., Ltd., Shanghai, China, D1771) with different concentrations of 0 μM, 5 μM, 10 μM, 20 μM, etc., in the above medium containing 5% FBS. Besides, cells in the control group (0 μM) were only intervened by the medium.
Cells were then inoculated into a six-well plate (2×105 cells/well) and left overnight. We applied Lipofectamine 2000 (Yanjin Biotechnology Co., Ltd., Shanghai, China, 11,668–019) for cell transfection. Transfectants included TINCR over-expression vector (TINCR) and corresponding control (NC), TINCR inhibition vector (si-TINCR) and corresponding control (si-NC), miR-761 mimics (miR-761) and corresponding control (miR-NC), and miR-761 inhibitor (anti-miR-761) and corresponding control (anti-miR-NC).
RT-PCR
The relative level of TINCR was quantified by RT-PCR and standardized by 3-phosphoglyceraldehyde dehydrogenase (GAPDH). At first, we used TRIzol (Baixu Biotechnology Co., Ltd., Beijing, China, RP11, 100 mL) to extract RNA from TNBC or BC serum samples and TNBC cells. Then, M-MLV reverse transcriptase kit (Yubo Biological Technology Co., Ltd., Shanghai, China, YB11105ES90) was utilized to synthesize cDNA. The relative mRNA expression was normalized by 2−ΔΔCT method. RT-PCR was performed using Applied Biosystems 2720 (Haonuosi Technology Co., Ltd., Beijing, China, 2,410,241,916).
Western Blot Analysis
TNBC cells were inoculated into a six-well plate (2×105 cells/well) and incubated overnight. The cells were washed twice with ice-cold phosphate buffer (PBS) (Zhenyu Biotechnology Co., Ltd., Shanghai, China, IH0019) and scraped on ice. Cells were lysed by lysis buffer (Shanghai China Wine Da Industrial Co., Ltd., Shanghai, China, N653-100ML) and incubated at 4°C for an hour. Then, the lysate was centrifuged at 10,000×g for 30 min at 4°C to collect the supernatant. Equal amount of protein (50 μg) was subjected to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Xinfan Biotechnology Co., Ltd., Shanghai, China, XFB1005), and then transferred to polyvinylidene fluoride (PVDF) membrane (Yanhui Biotechnology Co., Ltd., Shanghai, China, BSP0161). The membrane was incubated with blocking buffer (MineBio Life Sciences Ltd., Shanghai, China, BF01-1L) at room temperature for an hour, then incubated with primary antibodies of E-cadherin, N-cadherin and vimentin at 4°C overnight, and washed by TBST (Jichun Industrial Co., Ltd., Shanghai, China, JC-A80256). Then, the membrane was incubated with goat anti-mouse IgG secondary antibody (Lianke Biotechnology Co., Ltd., Shanghai, China, GAM007&ST001) bound with HRP for an hour at room temperature. After TBST washing again, the protein level was quantitatively analyzed by ECL solution (Yiji Industries Co., Ltd., Shanghai, China, YIJI12045.1) and X-ray film (Baiaolaibo Technology Co., Ltd., Beijing, China, QN3155-NGY).
Cell Migration and Invasion Determination
Cells were inoculated into serum-free medium (ZEPING Bioscience & Technologies Co., Ltd., Beijing, China, M293TIS) in the upper compartment for cell migration analysis, or inoculated into non serum-free medium (Fusheng Industrial Co., LTD., Shanghai, China, FS-79,049) that was precoated with Matrigel on the bottom for cell invasion analysis. The lower compartment was filled with complete medium (Cyagen Biosciences Inc., Suzhou, China, NKSFM-9006c). After incubation, the cells were fixed with methanol for 10 min, then stained with 0.5% crystal violet (Mairuibo Biotechnology Co., Ltd., Beijing, China, M DZ0054) and counted under a microscope.
Cell Proliferation Determination
Cell counting kit 8 (CCK-8) (Taize Jiaye Technology Development Co., Ltd., Beijing, China, MA0218) was used for analysis, and 2×103 cells were inoculated into 96-well plate. Cells were treated with different concentrations of LU for 24 hours, after which cells were collected and washed with PBS. The proliferation ability of cells cultured for 0, 24, 48 and 72 hours was detected by CCK-8 assay, and the cell growth curve was drawn based on the absorbance value at each time point.
Cell Apoptosis Determination
The percentage of apoptotic cells was detected by annexin V- fluorescein isothiocyanate (FITC) apoptosis detection kit (Yanhui Biotechnology Co., Ltd., Shanghai, China, 640,914). TNBC cells were inoculated in a 6-well plate (2×105 cells/well) and incubated overnight. Cells were treated with various concentrations of LU for 24 hours, after which cells were collected and washed with PBS. After centrifugation at 1500Xg for 10 min at 4°C, the precipitated cells were resuspended in annexin binding buffer. After that, the cells were stained by annexin V-FITC and propidium iodide (PI). The Annexin V+/PI- staining showed early apoptosis, and Annexin V+/PI+staining showed late apoptosis. Finally, BD FACS Canto II flow cytometry (FCM) (Genes Tech Technology Co., Ltd., Shanghai, China, BD001) and CellQuest Pro software were used to quantitatively analyze the apoptosis level.
Dual Luciferase Reporter
Wild type and mutant type (TINCR-wt, TINCR-Mut) of TINCR were co-transfected with miR-761 and miR-NC into TNBC cells. After transfection for 48 hours, the luciferase activity was measured by dual luciferase gene detection kit (Kanglang Biological Technology Co., Ltd., Shanghai, China, KL-D5868), and the detection steps were conducted strictly in accordance with the manual. The relative activity of firefly luciferase was standardized to the activity of renilla luciferase.
RNA Pull-Down Assay
In RNA pull-down assay, we incubated biotinylated TINCR with TNBC cell protein extract, and then evaluated the level of miR-761 by RT-PCR.
Statistical Analysis
We analyzed data and generated pictures by SPSS version 22.0. All experimental results were expressed as mean±SD. The differences of data results were analyzed by T-test or ANOVA. In all cases, P<0.05 was defined as statistically significant.
Results
TINCR and miR-761 in Serum of Early TNBC Patients are Significantly Up-Regulated
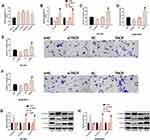
We explored the potential clinical application of TINCR and miR-761. First of all, we found that TINCR and miR-761 were notably up-regulated in the serum of early TNBC patients when compared with early BC patients (Figure 1A and B), suggesting that the two may participate in the pathological development of early TNBC. Furthermore, Pearson correlation coefficient analysis showed a significant positive correlation between the two (r=0.646, P<0.001) (Figure 1C), indicating that they might play a synergistic role in the pathological development of TNBC. Then, the receiver operating characteristic (ROC) curves of TNBC and BC were drawn, and the area under the curve (AUC) of TNBC and BC were 0.868 and 0.865, respectively (Figure 1D), which indicated that they had good screening value for TNBC. More details are shown in Figure 1.
TINCR Can Regulate the Metastatic Potential of TNBC Cells
We analyzed the effects of TINCR on the metastatic potential of TNBC cells through cell function experiments. TINCR was remarkably up-regulated in TNBC cell lines (Figure 2A), and its expression was higher in BT-549 and SUM159PT cells. Therefore, the two cells were chosen for later analysis. Then, we transfected TINCR and si-TINCR to achieve up-regulation and down-regulation of TINCR, respectively (Figure 2B). Furthermore, we found that down-regulation of TINCR remarkably inhibited the migration (Figure 2C and D), invasion (Figure 2E and F), and EMT (Figure 2G and H) of BT-549 and SUM159PT cells, and the inhibition of EMT showed a significant increase in epithelial marker (E-cadherin) and a significant decrease in mesenchymal marker (N-cadherin, vimentin). On the other hand, we found that up-regulation of TINCR notably increased the above metastatic potential of BT-549 and SUM159PT cells. Taken together, TINCR can regulate the metastatic potential of TNBC cells. More details are shown in Figure 2.
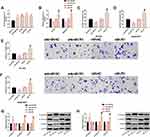
MiR-761 Can Also Regulate the Metastatic Potential of TNBC Cells
We also verified the potential regulation of miR-761 on the metastatic potential of TNBC cells. First, miR-761 expression in TNBC cells was found to be generally high (Figure 3A), and the expression level was more significant in BT-549 and SUM159PT cells. Therefore, these two cells were chosen for follow-up study. Then, we transfected miR-761 and anti-miR-761 into these two kinds of cells to achieve miR-761 over-expression and low-expression, respectively (Figure 3B), and found that miR-761 could regulate the metastatic potential of TNBC cells. Besides, miR-761 up-regulation could stimulate the metastatic potential of TNBC cells, while its down-regulation could notably inhibit the metastatic potential of TNBC cells. Our evaluation of the metastatic potential of cells was mainly based on cell migration (Figure 3C and D), invasion (Figure 3E and F) and EMT (Figure 3G and H). More details are shown in Figure 3.
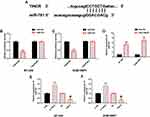
TINCR Acts as a Molecular Sponge for miR-761
We further explored the potential mechanisms of TINCR and miR-761, and verified the relationship between them through biological analysis. At first, they were found to have binding sites in miRDB (Figure 4A). According to dual luciferase reporter, compared with TINCR-Mut, miR-761 mimics considerably reduced luciferase activity via binding with TINCR-Wt (Figure 4B and C). Compared with Bio-NC, Bio-TINCR could recruit miR-761 in RNA pull-down test (Figure 4D). In addition, RT-PCR revealed that TINCR up-regulation could lead to marked miR-761 increase in TNBC cells, and TINCR down-regulation could lead to marked miR-761 decrease (Figure 4E and F). All the results pointed out that TINCR can target and regulate the expression of miR-761. More details are shown in Figure 4.
TINCR-miR-761 Molecular Module Has Certain Influence on the Anti-TNBC Activity of LU
We explored the role of TINCR-miR-761 molecular module in the anti-TNBC activity of LU. At first, we analyzed whether LU was related to the molecular module of TINCR-miR-761, and the results showed that LU had dose-dependent negative regulatory effects on TINCR and miR-761 (Figure 5A and B). Then, we explored the anti-TNBC activity of LU (10 μM) in cell proliferation and apoptosis. The results showed that LU could notably inhibit the proliferation of BT-549 cells (Figure 5C) and induce apoptosis (Figure 5E), and these effects were partially weakened by the abnormal up-regulation of TINCR or miR-761 (Figure 5C and E). In addition, the effects of miR-761 on the anti-TNBC activity of LU could be offset under the action of si-TINCR (Figure 5D and F). The above results enlightened us that the molecular module of TINCR-miR-761 can regulate the anti-TNBC activity of LU. More details are shown in Figure 5.
Discussion
In this study, we found that compared with the early BC patients, both TINCR and miR-761 were significantly up-regulated in serum samples of patients with early TNBC, suggesting that they might respond to the pathological mechanism of TNBC. TNBC is a severe subtype of BC, which has poor prognosis due to the lack of targeted therapy.15 Therefore, exploring the related molecular mechanism of TNBC is of great value for further elucidating the pathogenesis and finding effective therapeutic targets.
More and more scholars are focusing on the role of TINCR and miR-761 in tumors. For example, in the study of Xu et al.16 TINCR is notably up-regulated in gastric cancer (GC), and can change the viability of tumor cells by affecting staufen1 and KLF2. As reported by Han et al.17 TINCR is highly expressed in bladder urothelial carcinoma, and depletion of this gene inhibits the survival and motor ability of tumor cells. The expression of TINCR increases remarkably in BC trastuzumab resistant cells, and knocking down its expression can help improve drug resistance and EMT.18 All the above studies proved that TINCR tends to exhibit carcinogenic activity in various tumors and has carcinogenic effect. As for miR-761, it is also at a high level in non-small cell lung cancer (NSCLC)19, GC,20 TNBC and other tumors,14 and has certain carcinogenicity. In our clinical study, the correlation between TINCR and miR-761 was also analyzed, and a significant positive correlation between the expression of the two in TNBC serum samples was revealed (r=0.646, P<0.001). Besides, we found that the above two genes had high value in distinguishing early BC from TNBC, and their AUC were 0.868 and 0.865, respectively, both of which are not lower than 0.850, suggesting high value in distinguishing BC from TNBC.
In our cell research, TINCR and miR-761 were over-expressed in TNBC cell lines, suggesting that they may mediate the molecular mechanism of TNBC. We changed the expression of these two genes by cell transfection, and found that up-regulation of TINCR could promote the migration, invasion and EMT of TNBC cells, while down-regulation of TINCR led to significant opposite results. Such results indicated that TINCR can enhance the metastatic potential of TNBC cells, and the preparation of TINCR biological inhibitors may be beneficial to the treatment of TNBC patients by inhibiting the metastatic potential of TNBC cells. In the report of Liu et al.21 TINCR has a similar effect in BC cells, that is, its up-regulation can remarkably stimulate the migration and invasion of BC cells, while silencing TINCR can inhibit the mobility of BC cells. We also conducted similar research on miR-761. Fortunately, we obtained a result similar to that of TINCR, that is, miR-761 over-expression could enhance the tumor metastasis of TNBC, and its down-regulation could inhibit this tumor metastasis. Guo et al14 pointed out that miR-761 is an oncogene of TNBC. Its up-regulation can improve the survival and metastasis potential of tumor cells, while its down-regulation can suppress the malignant performance of tumor, which is similar to our research results.
Here, we confirmed a targeted relationship between TINCR and miR-761 through dual luciferase gene report and RNA pull-down experiment, and found that the former could actively regulate the expression of the latter. Zheng et al22 also confirmed that there is a target-controlled relationship between them. TINCR can be regarded as a competitive endogenous RNA of miR-761, which positively controlled the expression of miR-761 to regulate the migration of mesenchymal stem cells. One study suggested that both GAS5 and miR-137 reduced in melanoma tissues, and GAS5 positively regulates miR-137 in a targeted manner, which has certain similarities with our research.23 MiR-761 also has numerous targets, which are controlled by lncRNA in a variety of diseases to mediate the disease process. For example, miR-761 can mediate the development of NSCLC through the regulation of FOXF1 adjacent non-coding developmental regulatory RNA (FENDRR),24 participate in the development of thyroid papillary carcinoma through the target control of HOXA11 antisense RNA (HOXA11-AS),25 and is targeted by Arrl1 to regulate neurite growth during neurite regeneration in rats.26 However, only the molecular module of TINCR-miR-761 is highly correlated with TNBC, which has been confirmed in our research. Finally, we also studied the influence of TINCR-miR-761 molecular module on the anti-TNBC mechanism of LU. The results showed that LU could down-regulate the expression levels of TINCR and miR-761 in a dose-dependent manner, and could inhibit the proliferation and induce apoptosis of tumor cells in TNBC. This protective effect, however, was partially weakened when TINCR and miR-761 were abnormally up-regulated. Furthermore, the effect of miR-761 on the anti-TNBC effect of LU was partially eliminated under the intervention of si-TINCR. The above results indicate that the abnormality of the molecular module of TINCR-miR-761 can partially offset the anti-TNBC effect of LU.
The novelty of this study lies in the discovery for the first time that the molecular module TINCR-miR-761 can specifically respond to early TNBC, which has serum diagnostic value, and the abnormality of this molecular module is also related to the metastatic potential of TNBC cells and the anti-tumor activity of LU. Despite the above results, there is still room for improvement. First of all, we can analyze the underlying relationship between the molecular module and the clinicopathological parameters of TNBC patients, which has certain value for supplementing the clinical application of the molecular module. Secondly, we can supplement in vivo experiment of this molecular module in TNBC and the basic research exploration of drug resistance mechanism, which is beneficial to providing new cognition of TNBC in treatment. We will gradually improve this research in the future research.
Conclusion
To sum up, the imbalance of the molecular module of TINCR-miR-761 promotes the metastatic potential of early TNBC and partially counteracts the anti-tumor activity of LU, which may provide a new reference for the treatment of TNBC patients.
Funding
The authors received no funding for this work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Naderi N, Peymani M, Ghaedi K. The protective role of rs56103835 against breast cancer onset in the Iranian population. Mol Genet Genomic Med. 2019;7(3):e540. doi:10.1002/mgg3.540
2. Sharma R. Breast cancer incidence, mortality and mortality-to-incidence ratio (MIR) are associated with human development, 1990–2016: evidence from global burden of disease study 2016. Breast Cancer. 2019;26(4):428–445. doi:10.1007/s12282-018-00941-4
3. Beckers RK, Selinger CI, Vilain R, et al. Programmed death ligand 1 expression in triple-negative breast cancer is associated with tumour-infiltrating lymphocytes and improved outcome. Histopathology. 2016;69(1):25–34. doi:10.1111/his.12904
4. Lee J, Lim B, Pearson T, et al. Anti-tumor and anti-metastasis efficacy of E6201, a MEK1 inhibitor, in preclinical models of triple-negative breast cancer. Breast Cancer Res Treat. 2019;175(2):339–351. doi:10.1007/s10549-019-05166-3
5. Zhang G, Li H, Sun R, et al. Long non-coding RNA ZEB2-AS1 promotes the proliferation, metastasis and epithelial mesenchymal transition in triple-negative breast cancer by epigenetically activating ZEB2. J Cell Mol Med. 2019;23(5):3271–3279. doi:10.1111/jcmm.14213
6. Lin D, Kuang G, Wan J, et al. Luteolin suppresses the metastasis of triple-negative breast cancer by reversing epithelial-to-mesenchymal transition via downregulation of beta-catenin expression. Oncol Rep. 2017;37(2):895–902. doi:10.3892/or.2016.5311
7. Zhang G, Lan Y, Xie A, et al. Comprehensive analysis of long noncoding RNA (lncRNA)-chromatin interactions reveals lncRNA functions dependent on binding diverse regulatory elements. J Biol Chem. 2019;294(43):15613–15622. doi:10.1074/jbc.RA119.008732
8. Wong L, Huang YA, You ZH, Chen ZH, Cao MY. LNRLMI linear neighbour representation for predicting lncRNA-miRNA interactions. J Cell Mol Med. 2020;24(1):79–87. doi:10.1111/jcmm.14583
9. Wang J, Ding Y, Wu Y, Wang X. Identification of the complex regulatory relationships related to gastric cancer from lncRNA-miRNA-mRNA network. J Cell Biochem. 2020;121(1):876–887. doi:10.1002/jcb.29332
10. Jin C, Yan B, Lu Q, Lin Y, Ma L. Reciprocal regulation of Hsa-miR-1 and long noncoding RNA MALAT1 promotes triple-negative breast cancer development. Tumour Biol. 2016;37(6):7383–7394. doi:10.1007/s13277-015-4605-6
11. Li S, Zhou J, Wang Z, Wang P, Gao X, Wang Y. Long noncoding RNA GAS5 suppresses triple negative breast cancer progression through inhibition of proliferation and invasion by competitively binding miR-196a-5p. Biomed Pharmacother. 2018;104:451–457. doi:10.1016/j.biopha.2018.05.056
12. Xu S, Kong D, Chen Q, Ping Y, Pang D. Oncogenic long noncoding RNA landscape in breast cancer. Mol Cancer. 2017;16(1):129. doi:10.1186/s12943-017-0696-6
13. Abhishek S, Palamadai Krishnan S. Epidermal differentiation complex: a review on its epigenetic regulation and potential drug targets. Cell J. 2016;18(1):1–6. doi:10.22074/cellj.2016.3980
14. Guo GC, Wang JX, Han ML, Zhang LP, Li L. microRNA-761 induces aggressive phenotypes in triple-negative breast cancer cells by repressing TRIM29 expression. Cell Oncol (Dordr). 2017;40(2):157–166. doi:10.1007/s13402-016-0312-6
15. Jagadish N, Devi S, Gupta N, Suri V, Suri A. Knockdown of A-kinase anchor protein 4 inhibits proliferation of triple-negative breast cancer cells in vitro and in vivo. Tumour Biol. 2020;42(4):1010428320914477. doi:10.1177/1010428320914477
16. Xu TP, Liu XX, Xia R, et al. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34(45):5648–5661. doi:10.1038/onc.2015.18
17. Han Y, Sun G. Overexpression of lncRNA TINCR is associated with high-grade, invasive, and recurring tumors, and facilitates proliferation in vitro and in vivo of urothelial carcinoma of the bladder. Urol Oncol. 2020;38(9):738 e731–738 e738. doi:10.1016/j.urolonc.2019.12.026
18. Dong H, Hu J, Zou K, et al. Activation of LncRNA TINCR by H3K27 acetylation promotes trastuzumab resistance and epithelial-mesenchymal transition by targeting MicroRNA-125b in breast cancer. Mol Cancer. 2019;18(1):3. doi:10.1186/s12943-018-0931-9
19. Yan A, Yang C, Chen Z, Li C, Cai L. MiR-761 promotes progression and metastasis of non-small cell lung cancer by targeting ING4 and TIMP2. Cell Physiol Biochem. 2015;37(1):55–66. doi:10.1159/000430333
20. Sun X, Hou H, Li K, Zheng M. microRNA-761 regulates glycogen synthase kinase 3beta expression and promotes the proliferation and cell cycle of human gastric cancer cells. Oncol Lett. 2018;16(3):3459–3464. doi:10.3892/ol.2018.9133
21. Liu Y, Du Y, Hu X, Zhao L, Xia W. Up-regulation of ceRNA TINCR by SP1 contributes to tumorigenesis in breast cancer. BMC Cancer. 2018;18(1):367. doi:10.1186/s12885-018-4255-3
22. Zheng J, Huang Y, Li Y, et al. lncRNA-TINCR functions as a competitive endogenous RNA to regulate the migration of mesenchymal stem cells by sponging miR-761. Biomed Res Int. 2020;2020:9578730.
23. Bian D, Shi W, Shao Y, Li P, Song G. Long non-coding RNA GAS5 inhibits tumorigenesis via miR-137 in melanoma. Am J Transl Res. 2017;9(3):1509–1520.
24. Zhang G, Wang Q, Zhang X, Ding Z, Liu R. LncRNA FENDRR suppresses the progression of NSCLC via regulating miR-761/TIMP2 axis. Biomed Pharmacother. 2019;118:109309. doi:10.1016/j.biopha.2019.109309
25. Yin X, Zhang J, Li C, et al. LncRNA HOXA11-AS accumulation-induced microRNA-761 downregulation regulates cell growth by targeting TRIM29 in papillary thyroid cancer. Am J Transl Res. 2019;11(11):6826–6837.
26. Wang D, Chen Y, Liu M, et al. The long noncoding RNA Arrl1 inhibits neurite outgrowth by functioning as a competing endogenous RNA during neuronal regeneration in rats. J Biol Chem. 2020;295(25):8374–8386. doi:10.1074/jbc.RA119.011917
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.