Back to Journals » Journal of Asthma and Allergy » Volume 15
IL-13 Regulates Orai1 Expression in Human Bronchial Smooth Muscle Cells and Airway Remodeling in Asthma Mice Model via LncRNA H19
Authors Xiang LL, Wan QQ, Wang YM , He SJ, Xu WJ, Ding M, Zhang JJ, Sun YL, Dong X, Zhou Y, Cui YB , Gao YD
Received 28 January 2022
Accepted for publication 20 August 2022
Published 7 September 2022 Volume 2022:15 Pages 1245—1261
DOI https://doi.org/10.2147/JAA.S360381
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Amrita Dosanjh
Lin-Li Xiang,1,* Qian-Qian Wan,2,* Yi-Min Wang,1 Shao-Jun He,3 Wen-Juan Xu,3 Mei Ding,1,4 Jin-Jin Zhang,1,4 Yuan-Li Sun,1,4 Xiang Dong,1,4 Ying Zhou,5 Yu-Bao Cui,6 Ya-Dong Gao1,4
1Department of Allergology, Zhongnan Hospital of Wuhan University, Wuhan, People’s Republic of China; 2Department of Rheumatology, Zhongnan Hospital of Wuhan University, Wuhan, People’s Republic of China; 3Department of Respiratory and Critical Care Medicine Zhongnan Hospital of Wuhan University, Wuhan, People’s Republic of China; 4Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan, People’s Republic of China; 5Department of Pediatrics Laboratory, The Affiliated Wuxi Children’s Hospital of Nanjing Medical University, Wuxi, People’s Republic of China; 6Department of Clinical Laboratory, The Affiliated Wuxi People’s Hospital of Nanjing Medical University, Wuxi, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ya-Dong Gao, Department of Allergology, Zhongnan Hospital of Wuhan University, Donoghue Road 169, Wuhan, People’s Republic of China, Email [email protected]
Background: Increased proliferation and hypertrophy of airway smooth muscle cells (ASMCs) contribute substantially to airway remodeling in asthma. Interleukin (IL)-13 regulates ASMC proliferation by increasing Orai1 expression, the pore-forming subunit of store-operated Ca2+ entry (SOCE). The underlying mechanisms of this effect are not fully understood.
Methods: Bioinformatic analysis identified an interaction between microRNA 93– 5p (miR-93-5p) and long non-coding RNA (lncRNA) H19, and between miR-93-5p and Orai1. RNA interference was used to investigate H19 knockdown on IL-13-induced proliferation and migration of in vitro cultured human bronchial smooth muscle cells (hBSMCs). Functional relevance of H19 in airway inflammation and airway remodeling was investigated in murine models of acute and chronic asthma.
Results: IL-13 concentration-dependently increased the expression of H19 and Orai1 and decreased the expression of miR-93-5p in hBSMCs. H19 knockdown partly reversed the effects of IL-13 on the expression of miR-93-5p and Orai1 and attenuated the proliferation and migration of hBSMCs promoted by IL-13. IL-13-promoted expression of Orai1 was attenuated by miR-93-5p mimic and increased by miR-93-5p inhibitor. IL-13-promoted proliferation of hBSMCs was increased by miR-93-5p inhibitor but not affected by miR-93-5p mimic, whereas IL-13-promoted migration of hBSMCs was increased by miR-93-5p inhibitor and attenuated by miR-93-5p mimic. The inhibiting effect of H19 knockdown on IL-13-induced Orai1 expression and the proliferation and migration of hBSMCs was counteracted by miR-93-5p inhibitor but only marginally or not impacted by miR-93-5p mimic. The expression of H19 and Orai1 was higher in the lungs of asthmatic mice than in control mice. In asthmatic mice, H19 siRNA reduced Orai1 expression, inflammatory cell infiltration, goblet cell hyperplasia, collagen deposition and smooth muscle mass in the lungs.
Conclusion: H19 may mediate the effects of IL-13 on Orai1 expression by inhibition of miR-93-5p in hBSMCs. H19 may be a therapeutic target for airway inflammation and airway remodeling.
Keywords: IL-13, Orai1, lncRNA H19, microRNA-93-5p, bronchial smooth muscle cells, asthma model
Background
Asthma is a heterogeneous disease characterized by variable respiratory symptoms and airflow limitation.1 Type 2 inflammation is now believed to be the immune mechanism responsible for most cases of allergic asthma.2 IL-13 is a type 2 cytokine that can be produced by T helper 2 (Th2) cells and group 2 innate lymphoid cells (ILC2).2
Airway remodeling is an important feature of asthma that causes persistent airflow limitation and is associated with decreased reversibility. Airway remodeling may contribute to steroid resistance in asthma patients with persistent symptoms.3 The presence of airway remodeling in early childhood with asthma suggests the dissociation between airway inflammation and airway remodeling.1 Pathological changes associated with airway remodeling include epithelial damage, cilial dysfunction, goblet cell hyperplasia, thickened lamina reticularis and reticular basement membrane, increased subepithelial collagen deposition and angiogenesis, and airway smooth muscle hypertrophy and hyperplasia.1 Airway smooth muscle cell (ASMC) hypertrophy and hyperplasia represent the major components of airway remodeling, and ASM mass has been regarded as the best predictor for airflow limitation in asthma.1
IL-13 plays a pivotal role in airway hyperreactivity (AHR)4 and airway remodeling.2 It acts on different cell types to induce airway remodeling, including airway epithelial cell,5 fibroblasts,6 and more importantly, ASMCs.7 We demonstrated previously that store-operated Ca2+ entry (SOCE) contributes to airway inflammation and remodeling in asthma.8 IL-13 induces ASMC proliferation by promoting Ca2+ release and SOCE.9 Orai1 is the pore-forming subunit of store-operated Ca2+ (SOC) channels and mediates Ca2+ influx upon interaction with stromal interacting molecule 1 (STIM1). STIM1 senses the Ca2+ level in the endoplasmic reticulum (ER) Ca2+ store and translocates to ER membrane underneath plasma membrane after Ca2+ store depletion, where it interacts with Orai1 and activates SOCE.10 Upregulated expression of Orai1 was shown in proliferating ASMCs and ASMCs from asthmatic mice, whereas knockdown of Orai1 attenuated SOCE and ASMC proliferation.11,12 Caveolae also regulate SOCE in ASMCs via increasing the expression of Orai1.13 All these results suggest a critical role of Orai1 in the functional regulation of ASMCs in the context of asthma. However, the mechanisms underlying the regulating effects of IL-13 on Orai1 in ASMCs are still not fully elucidated.
Long non-coding RNAs (lncRNAs) are non-coding RNAs of at least 200 nucleotides.14 Three major functional principles are assigned to lncRNAs: RNA-based function, regulatory element in the gene body and transcription-based function.15 LncRNAs have been shown to participate in the pathogenesis of airway inflammation and airway remodeling. LncRNAs may act as biomarkers for asthma phenotypes and also for glucocorticoids sensitivity in asthma.16 A few lncRNAs have been proved to be involved in the airway remodeling by acting on ASMCs.16,17 For instance, growth arrest specific-5 (GAS5), TCF7, TUG1, Brain cytoplasmic RNA 1 (BCYRN1), LINC00882, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), and COPDA1 promoted proliferation of ASMC, whereas plasmacytoma variant translocation (PVT1) inhibited the growth of ASMC.16,17
We aimed to determine the potential lncRNAs that regulate the expression and function of Orai1. By using bioinformatic analysis, we first identified several microRNAs that interact with Orai1. Among them, microRNA (miRNA) has-miR-93-5p (miR-93-5p) has been demonstrated to participate in gene expression in asthma.18 We then identified a few lncRNAs that interact with miR-93-5p. Of them, lncRNA H19 plays a critical role in diverse biological and pathophysiological processes including tumor metastasis and progression, hypoxia, metabolism, oxidative stress and inflammation.19 The role of H19 in airway inflammation and ASMCs is still unknown. It is reported that H19 promotes pulmonary smooth muscle cell proliferation by sponging miRNA let-7b.19 Since LncRNA/miRNA/mRNA axis is the major mechanism of lncRNA regulating biological processes,20 we investigated the potential roles of H19 in the regulation of Orai1 expression, ASMC proliferation and migration under stimulation with IL-13. In addition, the role of H19 in the regulation of airway inflammation and airway remodeling was also examined in murine asthma models.
Materials and Methods
Bioinformatic Analysis
LncRNAs regulate mRNA expression by sponging microRNAs. We first determined to find the microRNAs potentially interact with Orai1. By searching on the public websites TargetScan (http://www.targetscan.org) and ENCORI (http://starbase.sysu.edu.cn), we identified eight microRNAs in TargetScan and one hundred microRNAs in ENCORI that target human Orai1 (Gene ID: 84876). The eight microRNAs identified in TargetScan were also included in those microRNAs identified in ENCORI: hsa-miR-20a-5p, hsa-miR-519d-3p, hsa-miR-526b-3p, hsa-miR-106b-5p, hsa-miR-93-5p, hsa-miR-106a-5p, hsa-miR-20b-5p, and hsa-miR-17-5p. By searching in Pubmed, we found that microRNA-93 has been reported to regulate the expression of Orai1 in renal fibrosis.21 By using DIANA tools (http://diana.imis.athena-innovation.gr/DianaTools/index.php) ten LncRNAs that target hsa-miR-93-5p were identified: AC021078.1, SNHG16, MALAT1, XIST, ARHGAP27P1-BPTFP1-KPNA2P3, AC004656.1, AC021092.1, AC243964.3, AC005261.1 and H19. LncRNA H19 has been proven to play an important role in fibrotic diseases as well as inflammatory processes.2,22,23 Thus, H19 was selected as the candidate lncRNA of this study.
In vitro Culture of hBSMCs
Segmental bronchi were obtained from lung cancer patients undergoing pulmonary lobectomy or pneumonectomy in Zhongnan Hospital of Wuhan University. Informed consent was acquired from all patients and the study was approved by Medical Ethics Committee of Zhongnan Hospital (Approval number: 2019044). Bronchial smooth muscle was dissected out and cut into small pieces, which were then placed into 25 cm2 culture flasks and cultured in RPMI-1640 medium containing L-glutamine (2 mM), penicillin (50 U/mL), and streptomycin (50 mg/mL), and supplemented with 10% fetal bovine serum (FBS). After reaching ~80% confluence, cells were passaged using tyrosination with 0.05% trypsin-EDTA and then grown in high glucose DMEM/F12 medium (Gibco-BRL, Carlsbad, CA, USA) supplemented with 20% FBS (Gibco-BRL). Typical hBSMCs exhibited a hill-and-valley pattern when reaching confluence. The purity of hBSMCs was confirmed by immunocytochemistry with mouse anti-human α-smooth muscle actin (SMA) antibody, and 95% of cells were positively stained with anti-α-SMA (Figure SA). hBSMCs before passage 5 were used for the experiments. In these experiments, hBSMCs were seeded at a density of 5×104 cells/mL and cultured in DMEM/F12 with 10% FBS for 24 h. Cells were treated differently according to the purpose of each experiment.
Transfection of hBSMCs
The expression of lncRNA H19 in hBSMCs was inhibited with transfection of siRNA against H19 (50 nM). Scrambled siRNA was used as the negative control (Qijing, Wuhan, China). SiRNAs were diluted in riboFECTTMCP reagents (Ribobio, Guangzhou, China). Sequences of H19 siRNA were as follows: sense 5’-CCAACAUCAAAGACACCAUTT-3’, antisense 5’-AUGGUGUCUUUGAUGUUGGTT-3’. The expression of miR-93-5p was upregulated by miR-93-5p mimic and downregulated by miR-93-5p inhibitor (Ribobio, Guangzhou, China) respectively. MiR-93-5p mimic or inhibitor was transfected into hBSMCs with riboFECTTMCP reagents. For transfection, hBSMCs were inoculated into 24-well plates with a density of 1×104 cells/well and transfection was conducted when cells grew to 50~60% confluence. The transfection complex was added into DMEM/F12 containing 10% FBS with a final volume of 500 μL. Control cells were treated only with same volume of PBS. The medium containing transfection complex was removed after 6 h, and cells were cultured in DMEM/F12 in the presence of IL-13 for an additional 24 − 72 h. The silencing efficiency of siRNA was examined with qRT-PCR.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA from lung tissues or in vitro cultured hBSMCs was extracted with TRIzol reagents (Invitrogen, CA, USA) according to the manufacturer’s instructions. For Orai1 and H19, RNA was reversely transcribed into cDNA using a ReverTra Ace qPCR RT kit (Toyobo, Tokyo, Japan). cDNA was then amplified with SYBR Premix Ex Taq™ II (RR820A, Takara, Japan) via the following three steps: 1 cycle at 95°C for 30s, followed by 40 cycles of 95°C for 5s and 60°C for 30s, and a melting curve collected at 95°C for 5 s and 60°C for 1 min. For miR-93-5p detection, reverse transcription and qRT-PCR were performed using the Bulge-LoopTM miRNA qRT-PCR starter kit (R11067.2, RiboBio Co., Guangzhou, China) and the Bulge-LoopTM hsa-miR-93-5p qRT-PCR primer set (R10031.7, RiboBio Co., Guangzhou, China). The amplification process included 1 cycle at 95°C for 10 min, 40 cycles of 95°C for 2 s, 60°C for 20s, and 70°C for 10s. The cycle threshold (CT) values of miR-93-5p were normalized to U6, and CT values of H19 or Orai1 were normalized to β-actin for respectively. The reaction was performed using the CFX96 Real-Time system (Bio-Rad, USA). Data were analyzed using 2−ΔΔCT method.
Western-Blot
Lung tissues or hBSMCs were lysed in RIPA buffer containing phosphatase and protease inhibitors (ST505, Beyotime, China) on ice for 30 min and centrifuged at 12,000 rpm for 20 min at 4°C. Protein concentrations were detected using a BCA protein assay kit (Beyotime, China), and equal amounts of protein (40 μg each lane) were separated with SDS-PAGE and then transferred to Poly (vinylidene fluoride) (PVDF) membranes (Millipore, USA). Membranes were incubated with primary antibody of Orai1 (rabbit anti-human, 1:4000, Abcam, USA) and β-actin (goat anti-rabbit, 1:5000, Sigma, St. Louis, MO, USA) for 18 h, followed by horseradish peroxidase (HRP)-conjugated secondary antibodies for 2 h at room temperature. Enhanced chemiluminescence (ECL) was used to identify the immunoreactive bands, and blots densitometry was analyzed by ImageJ software.
CCK-8 Assay
The proliferation rates of hBSMCs were determined with the cell counting kit-8 (CCK-8) assay (Dojindo, Shanghai, China). hBSMCs were seeded in 96-wells plates at a concentration of 1×104 cells/well and transfected with siRNA against H19 or miR-93-5p mimic or miR-93-5p inhibitor. After 6 h, the medium containing transfection complexes was removed. Cells were then synchronized in FBS-free medium for 24 h, and then incubated in 1% FBS-containing medium in the presence of IL-13 (10 ng/mL) for 24 h or 48 h. 10 μL of CCK-8 reagent was added into each well and incubated for 2 − 4 h and the optical density (OD) values of absorbance at 450 nm were measured.
Scratch Wound Healing Assay
A scratch wound healing assay was conducted to determine the migration rates of hBSMCs. hBSMCs were cultured in 6-well plates in DMEM/F12 supplemented with 10% FBS until the cells reached 60% confluency. Cells were then transfected with siRNA against H19 or miR-93-5p mimic or inhibitor with the same process as in the CCK-8 assay. After removing transfection complexes and incubating for 24 h in a 10% FBS- containing medium, linear wound tracks were generated with sterile 1 mL pipettes and IL-13 (10 ng/mL) was added into each well. Non-adherent cells were removed by rinsing twice with sterile PBS before adding IL-13. DMEM/F12 with 1% FBS was used in this assay. The distance between the two edges of the wound was measured 24 h after scratching.
Dual-Luciferase Reporter Assay
The sequences of H19 and Orai1 3ʹUTR containing miR-93-5p binding sites were cloned into the downstream of pmirGLO luciferase reporter vector to form the wild-type (WT) constructs WT-H19 and Orai1 3ʹUTR-WT, respectively. The corresponding mutant (MUT) constructs MUT-H19 and Orai1 3ʹUTR-MUT were generated by mutating the miR-93-5p binding sites. For dual-luciferase reporter assay, 93T cells were co-transfected with these constructs and miR-93-5p mimic or control mimic using Lipofectamine™ 2000. After 48 h, cells were harvested and lysed for luciferase activity analysis with a dual-luciferase assay system.
Murine Models of Asthma
This study was approved by the Animal Ethics Committee of Zhongnan Hospital (Approval No. 20200708). BALB/c mice (6 − 8 weeks) were obtained from the Animal Experimental Center of Wuhan University and housed under pathogen-free conditions at a 12 h:12 h light: dark cycle. The construction of acute asthma mice model has been described previously.24 HDM extract (Stallergenes Greer, London, UK) resolved in PBS was intranasally instilled into the mice. In the 2-week acute asthma mice model, 100 µg HDM in 25 µL PBS was introduced to each mouse on day 1 − 5(Figure SBi), whereases the control mice were treated with an equal volume of PBS. To inhibit the expression of H19, mice of acute asthma model were treated with 50 μL lentivirus (1×108 TU/mL) carrying H19-specific siRNA or scramble siRNA and green fluorescence protein (GFP) gene (Genechem Co., LTD, Shanghai, China) (Figure SC) respectively by intranasal instillation on day 11 and 13, respectively. 4’,6-diamidino-2-phenylindole (DAPI) blue fluorescence stain was used to show cells’ location and transfection effect (Figure SC). Asthma mice models were challenged with 100 µg HDM in 25 µL PBS on day 12 and 14, and the control mice were treated with an equal volume of PBS.
For the 8-week chronic asthma model, mice were sensitized with 10 µg HDM in 35 µL PBS for 5 consecutive days in week 1. From week 2 to week 8, mice were challenged with the same concentration of HDM for five consecutive days per week. Control mice for chronic model were treated only with PBS in the same pattern. To inhibit the expression of H19, 1×108 TU of lentivirus carrying H19-specific siRNA or scramble siRNA and GFP gene was introduced into the chronic asthma mice by intranasal instillation on the day before the first HDM challenge every week from week 2 to week 8. Evaluation of the endpoint metrics was performed 24 h after the last instillation (Figure SBii).
Immunohistochemical Analysis
The left lung tissues of each mouse were fixed with 10% neutral formalin for 24 h, embedded in paraffin and sectioned at 4 mm for hematoxylin-eosin (HE) staining, periodic acid-Schiff (PAS) staining (Solarbio Technology, Beijing, China), Masson staining, and α-smooth muscle actin (α-SMA; 1:100, Abcam, USA) immunostaining to observe the pathological changes of lung tissues in mice.
Statistical Analysis
All data were generated from at least three independent assays and presented as mean ± standard deviation (SD). Student’s t-test was used to compare the difference between two groups and one-way ANOVA was used to compare the difference among three or more groups. All statistical analysis was performed with GraphPad Prism 8 software (La Jolla, CA, USA). P<0.05 was regarded as statistically significant.
Results
Interactions Among Orai1, lncRNA H19 and miR-93-5p
Bioinformatics predictions using TargetScan and ENCORI indicated that H19 and Orai1 sequences contain miR-93-5p binding sites (Figure SD). To determine the direct interactions among miR-93-5p, Orai1 3ʹUTR and H19, the miR-93-5p binding sites in Orai1 and H19 were mutated to generate Luc-Orai1-MUT and Luc-H19-MUT constructs respectively. Consistently, MiR-93-5p mimic, plasmid-mediated Orai1 overexpression construct (Luc-Orai1-WT) and H19 overexpression construct (Luc-H19-WT) were also generated for dual-luciferase reporter assay in 293T cells (Figure 1A and B). MiR-93-5p mimic inhibited the luciferase activity in Luc-H19-WT but not in Luc-H19-MUT (Figure 1C). Similarly, MiR-93-5p mimic inhibited the luciferase activity in Luc-Oria1-WT but not in Luc-Orai1-MUT (Figure 1D). These results confirmed that miR-93-5p interacts with both H19 and Orai1.
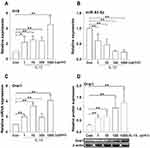
Effects of IL-13 on the Expression of H19, miR-93-5p and Orai1 in hBSMCs
The effects of IL-13 on the expression of H19 and miR-93-5p in in vitro cultured hBSMCs were determined with RT-PCR. IL-13 increased the expression of lncRNA H19 at all tested concentrations, maximally at 1000 ng/mL (Figure 2A). By contrast, IL-13 dose-dependently decreased the expression of miR-93-5p in hBSMCs (Figure 2B). Consistent with our previous results, IL-13 also increased both mRNA and protein expression of Orai1 in hBSMCs at all tested concentrations, with a maximal effect at 1000 ng/mL (Figure 2C and D). These results suggested that IL-13 upregulated the expression of H19 and Orai1 but inhibited the expression of miR-93-5p.
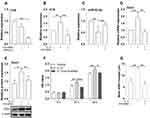
H19 Knockdown Inhibited the Expression of miR-93-5p and Orai1 in hBSMCs
We then determined whether H19 has a regulating effect on the expression of miR-93-5p and Orai1 induced by IL-13. qRT-PCR showed that H19-specific siRNA but not scrambled siRNA significantly decreased the expression of H19 in hBSMCs (Figure 3A). IL-13-promoted expression of H19 was inhibited by H19 knockdown with specific siRNA (Figure 3B). In addition, IL-13-inhibited expression of miR-93-5p was attenuated by H19 knockdown (Figure 3C). IL-13-promoted expression of Orai1, both the mRNA and protein, was inhibited by H19 knockdown in hBSMCs (Figure 3D and E). These results indicated that H19 mediated the effects of IL-13 on miR-93-5p and Orai1 expression in hBSMCs. The relation between H19, miR-93-5p and Orai1 were depicted in (Figure SE).
H19 siRNA Knockdown Inhibited the Proliferation and Migration of hBSMCs
The role of H19 in the proliferation and migration of hBSMCs induced by IL-13 was then investigated. As expected, IL-13 (10 ng/mL) significantly increased the proliferation of hBSMCs when compared with control cells. H19 knockdown significantly inhibited IL-13-stimulated proliferation of hBSMCs at both 24 h and 48 h (Figure 3F). Similarly, the migration of hBSMCs induced by IL-13 was also inhibited by H19 knockdown, as demonstrated in wound repair tests (Figure 3G). These results suggested that H19 was functionally relevant in IL-13-induced proliferation and migration of hBSMCs.
3.5 miR-93-5p Inhibited IL-13-Induced Orai1 Expression
Since miR-93-5p interacts with Orai1, and IL-13 had opposite effects on the expression of Orai1 and miR-93-5p, we then explored the role of miR-93-5p in IL-13-induced expression of Orai1 in hBSMCs by using miR-93-5p-specific mimic or inhibitor. Both the IL-13-promoted mRNA and protein expression of Orai1 were inhibited by miR-93-5p mimic and increased by miR-93-5p inhibitor (Figure 4A and B). These results suggested that miR-93-5p may counteract IL-13-induced Orai1 expression.
miR-93-5p Inhibited IL-13-Induced Proliferation and Migration of hBSMCs
We then determined the functional relevance of miR-93-5p in hBSMCs. miR-93-5p mimic had no significant effect on IL-13-induced proliferation of hBSMCs. However, IL-13-induced proliferation of hBSMCs was significantly increased by miR-93-5p inhibitor at both 24 h and 48 h (Figure 4C). By contrast, miR-93-5p mimic significantly attenuated, whereas miR-93-5p inhibitor significantly promoted IL-13-induced hBSMCs migration, as indicated with wound repair tests. (Figure 4D). These results indicated that miR-93-5p may partly reverse the effects ofIL-13 on the proliferation and migration of hBSMCs.
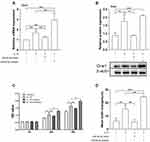
miR-93-5p Counteracted the Effect of H19 Knockdown on Orai1 Expression
To directly demonstrate the opposite effects of H19 and miR-93-5p on IL-13-induced Orai1 expression and biological functions in hBSMCs, we investigated the effects of miR-93-5p mimic and inhibitor on IL-13-induced Orai1 expression in hBSMCs with H19 knockdown. The inhibiting effect of H19 knockdown on IL-13-induced expression of Orai1 mRNA was amplified by miR-93-5p mimic and partly counteracted by miR-93-5p inhibitor (Figure 5A). The inhibiting effect of H19 knockdown on IL-13-induced expression of Orai1 protein was not significantly affected by miR-93-5p mimic but was significantly counteracted by miR-93-5p inhibitor (Figure 5B). These results indicate that miR-93-5p may inhibit the effect of IL-13 on Orai1 expression by counteracting the effect of H19 on hBSMCs.
miR-93-5p Counteracted the Promoting Effect of H19 on the Proliferation and Migration of hBSMCs
The functional relevance of the opposite effects of H19 and miRNA-93-5p was then determined in hBSMCs. H19 knockdown had a significant effect on the IL-13-induced proliferation of hBSMCs at 48 h but not 24 h. In the presence of IL-13 and H19 siRNA, the proliferation of hBSMCs was significantly inhibited by miR-93-5p mimic at 24 h but not at 48 h, whereas miR-93-5p inhibitor significantly increased the proliferation of hBSMCs at both 24 h and 48 h (Figure 5C). H19 knockdown significantly inhibited IL-13-induced hBSMCs migration, and this effect was significantly promoted by miR-93-5p inhibitor but not affected by miR-93-5p mimic (Figure 5D).
Role of H19 in Orai1 Expression and Airway Inflammation in the Murine Model of Acute Asthma
We then determined the in vivo role of lncRNA H19 in Orai1 expression and airway inflammation. HDM-sensitized acute asthma mice were transfected with lentivirus carrying H19-specific siRNA. The expression of H19 was elevated in the lungs of acute asthma mice compared with control mice. Transfection of H19-specific siRNA but not scrambled siRNA significantly inhibited the expression of H19 in the lungs of acute asthma mice (Figure 6A). Both the mRNA and protein expression of Orai1 in the lungs of acute asthma mice were significantly increased when compared with control mice (Figure 6B and C). Both the mRNA and protein expression of Orai1 in the lungs were inhibited by transfection with H19-specific siRNA but not scrambled siRNA (Figure 6B and C).
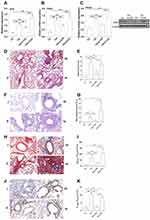
Substantial inflammatory cell infiltration was observed in peri-bronchial and perivascular areas of the lungs of acute asthma mice (Figure 6Dii) when compared with control mice (Figure 6Di). H19-specific siRNA (Figure 6Diii), but not scrambled siRNA (Figure 6Div), significantly reduced the inflammatory cell infiltration in the lungs of asthma mice. Inflammatory scores confirmed these results (Figure 6E). H19 siRNA knockdown also significantly inhibited bronchial goblet cell hyperplasia in acute asthma mice (Figure 6F), as quantified by the proportion of PAS+ area to perimeter of bronchi (Figure 6G). These results indicated that H19 may be involved in the pathogenesis of airway inflammation in asthma mice by increasing Orai1 expression.
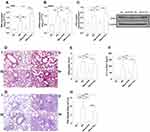
Role of H19 in Orai1 Expression, Airway Inflammation, and Airway Remodeling in the Chronic Asthma Murine Model
Consistent with that observed in acute asthma mice, the expression of H19 and the mRNA and protein expression of Orai1 were higher in the lungs of HDM-induced chronic asthma mice when compared with control mice (Figure 7A−C). Transfection of H19-specific siRNA but not scrambled siRNA significantly reduced the expression of H19 in the lungs of chronic asthma mice (Figure 7A). Transfection with H19-specific siRNA, but not scrambled siRNA, also inhibited both mRNA and protein expression of Orai1 in the lungs of chronic asthma mice (Figure 7B and C).
Substantial inflammatory cell infiltration was also observed in the lungs of chronic asthma mice when compared with control mice. H19-specific siRNA but not scramble siRNA reduced inflammatory cell infiltration in the lungs of chronic asthma mice (Figure 7D), as quantified with inflammatory scores (Figure 7E). PAS staining showed a prominent goblet cell hyperplasia in chronic asthma mice when compared with control mice. H19 siRNA but not scrambled siRNA inhibited goblet cell hyperplasia in bronchial mucosa (Figure 7F), as quantified by the ratio of PAS+ area to perimeter of bronchi (Figure 7G).
Masson staining showed a higher collagen deposition beneath bronchial mucosa in chronic asthma mice when compared with control mice. H19-specific siRNA but not scrambled siRNA significantly inhibited the collagen deposition in bronchi of chronic asthma mice (Figure 7H), as quantified by the ratio of the positive area of Masson staining to the area of peri-bronchial mucus (Figure 7I). Anti-α-SMA antibody staining showed a larger area of smooth muscle beneath the bronchial mucus in chronic asthma mice compared with control mice. H19-specific siRNA but not scrambled siRNA reduced the smooth muscle area in the lungs of chronic asthma mice (Figure 7J). The ratio of the positively stained area with α-SMA to that of the perimeter of bronchi confirmed these results (Figure 7K). Taken together, these data indicated that H19 may promote airway inflammation and airway remodeling by enhancing the expression of Orai1 in the lungs of chronic asthma mice.
Discussion
The dual-luciferase reporter assay confirmed the interactions between miR-93-5p and H19 as well as Orai1 in 293 T cells. We then proved these interactions in hBSMCs and found that IL-13 increased the expression of H19 and Orai1 and decreased the expression of miR-93-5p in hBSMCs, and this effect of IL-13 was inhibited by H19 knockdown. In addition, the promoting effect of IL-13 on the expression of Orai1 was inhibited by miR-93-5p. Moreover, the promoting effects of IL-13 on hBSMC proliferation and migration were attenuated by H19 knockdown and miR-93-5p mimic but were aggravated by miR-93-5p inhibitor. In asthma mice models, higher expression of H19 and Orai1 and more remarkable airway inflammation and remodeling were observed in the lungs of asthmatic mice when compared with control mice. H19 inhibition with siRNA attenuated not only the expression of Orai1, but also the airway inflammation and airway remodeling in the lungs of asthma mice.
Type 2 cytokine IL-13 can be produced by both Th2 cells and ILC2 cells.2 IL-13 is essential to the development of asthma25 by inducing different biological effects in inflammatory and structural cells of the airway.26 It has been shown that IL-13 contributes to airway remodeling of asthma by inducing airway epithelial barrier dysfunction,27,28 goblet cell metaplasia, mucus hypersecretion, ASMC proliferation and migration, fibroblasts proliferation,6 airway hyperreactivity and IgE production,26 all of which are features of airway remodeling. We have also previously shown that IL-13 could upregulate the expression of Orai1 and STIM1 and the activity of SOCE in cultured ASMCs.9 Consistently, the results in this study also showed that IL-13 dose-dependently increased the mRNA and protein expression of Orai1 in hBSMCs.
For the first time, we demonstrated that IL-13 increased the expression of lncRNA H19 and decreased the expression of miR-93-5p in hBSMCs. In addition to H19, previous studies also showed that other lncRNAs also play a role in IL-13-induced responses, eg, lncRNA BANCR in eosinophilic esophagitis29 and multiple lncRNAs in mycobacterium tuberculosis-infected macrophages.30 In allergic rhinitis, IL-13 decreased the expression of lncRNA Linc00632 in nasal epithelial cells, which then inhibited IL-13-induced production of GM-CSF, eotaxin and MUAC5AC.31
IL-13 was also shown to regulate the expression of multiple miRNAs associated with airway inflammation. In nasal epithelial cells, IL-13 promoted the expression of miR-49831 and dose-dependently decreased the expression of miR-15a-5p.32 In human bronchial epithelial cells, miR-330 inhibited IL-13-induced secretion of MUC5AC.33 miRNA microarray assay showed a higher expression of 79 miRNAs and a lower expression of 138 miRNAs in airway epithelial cells from asthmatic patients compared to that in healthy control subjects.34 Of note, among these miRNAs, the expression of miR-93-5p was lower in asthmatic patients. Moreover, the expression patterns of miRNAs were similar in the in vitro cultured human bronchial epithelial cells under stimulation with IL-13 and in asthmatic patients.34 We observed an inhibitory effect of IL-13 on the expression of miR-93-5p in hBSMCs. Similarly, IL-13 was reported to downregulate the expression of miR-133a35 and miR-140-3p36 in BSMCs, and resulted in the upregulated expression of RhoA, a possible key regulator of bronchial hyperreactivity. These data suggest that microRNAs, including miR-93-5p, may be actively involved in type 2 inflammation and airway remodeling in asthma. Further studies are warranted to investigate the expression of miR-93-5p in ASMCs from asthmatic patients.
H19 has been reported to interact with multiple miRNAs to regulate diverse biological processes. H19 was shown to upregulate the expression of angiotensin II type 1 receptor (AT1R) and promote the proliferation of pulmonary artery smooth muscle cell via interacting with miR-Let-7b.19 H19 was also shown to interact with miR-21 to regulate the proliferation and migration of ASMCs,37 and interact with miR-599 to promote the proliferation of vascular smooth muscle cells.38 In skeletal muscle, the only tissue with persistent H19 expression after birth, H19 was found to interact with miR-675-3p and miR-675-5p to promote the differentiation and regeneration of skeletal muscle.39 All these data, combined with our results of the interaction between H19 and miR-93-5p, support the conclusion that H19 may play an important role in the regulation of diverse biological functions mediated by miRNAs.
In addition to miR-93-5p, other microRNAs have also been reported to regulate the expression and biological activities of Orai1. MiR-93 was reported to downregulate the expression of Orai1 and suppress epithelial-mesenchymal transformation and fibrogenesis induced by transforming growth factor-β1 (TGF-β1).21 In addition, miR-519 could downregulate the expression of Orai1 and inhibit the growth of cancer cells.40,41 In CD4+ T cells, overexpression of miR-10a-5p decreased the expression of Orai1, the activity of SOCE, and the cell proliferation.42 Deletion of Dicer, the enzyme responsible for processing the precursor miRNAs to mature miRNAs, resulted in reduced Orai1 expression and SOCE in CD4+ T cells, indicating the importance of microRNAs both to the expression and to the function of Orai1.43 Together with our results, these data suggest that miRNAs are important to the proper function of Orai-mediated SOCE.
Functionally, the interactions between H19 and miRNAs have been proven to regulate the pathogenesis of several fibrotic diseases, eg, in pulmonary fibrosis by regulating the miR-196a/COL1A1,22 in renal fibrosis by regulating miR-29a2 and in cholestatic liver fibrosis by regulating the S1PR2/SphK2 and let-7/HMGA2 axis.23 The interactions between H19 and miRNAs were also reported to play a pivotal role in diverse inflammatory diseases. In rheumatoid arthritis, H19 promoted the inflammation by degradation of miR-103a.44 In ankylosing spondylitis, H19 interacted with miR-675-5p and miR-22-5p to increase the release of IL-17A/IL-23.45 In addition, H19 could regulate aging and age-related diseases by regulating inflammation.46 Re-expression of H19 in cancer tissues may contribute to the genesis and metastasis of different types of cancers, such as hepatocellular carcinoma,47 thyroid cancer cells48 and non-small cell lung cancer.49 H19 also has functional relevance in the regulation of hypoxia-induced tissue injury and oxidative distress.50 The role of H19 in the regulation of intracellular Ca2+ signaling is not clearly elucidated. Our results demonstrated a promoting effect of H19 on the expression of Orai1 and the proliferation and migration of hBSMCs. This is contradictory to another study showing that H19 inhibited platelet-derived growth factor (PDGF)-induced proliferation and migration of ASMCs.37 A lower expression of H19 was also found in ASMCs from patients with mild asthma compared to healthy controls.51 Therefore, further studies are warranted to clarify the precise role of H19 in the functional regulation of ASMCs.
Our study had several limitations. First, the role of H19 in the functional regulation of hBSMCs was investigated only with H19 siRNA but not with H19 overexpression. Second, the role of H19 in airway inflammation and airway remodeling needs to be confirmed in H19 knock-out mice. Finally, the activity of SOCE was not determined with Ca2+ fluorescence and/or electrophysiological measurements using whole-cell patch-clamp technique, due to technical unavailability. Further studies are warranted to investigate the effects and functional relevance of H19-miR-93-5p-Orai1 interactions on SOCE in ASMCs and other cell types that may contribute to the pathogenesis of asthma.
Conclusions
IL-13 can increase the proliferation and migration of airway smooth muscle cells by promoting the expression of H19 and Orai1 and decreasing the expression of miR-93-5p. H19 aggregates airway inflammation and airway remodeling in murine models of asthma possibly by inhibition of the expression of Orai1. H19 may be a novel target for the treatment of airway remodeling in asthma.
Abbreviations
α-SMA, α-smooth muscle actin; ASMCs, Airway smooth muscle cells; BALF, Bronchoalveolar lavage fluid; DMEM, Dulbecco’s modified eagle medium; FBS, fetal bovine serum; GFP, Green fluorescent protein; hBSMCs, human bronchial smooth muscle cells; HDM, House dust mites; PBS, Phosphate buffered solution; qRT-PCR, Quantitative Real-time PCR; IL-13, Interleukin-13; ILC2, Group 2 innate lymphoid cells; lncRNA, long non-coding RNAs; miRNA, microRNA; Orai1, CRACM1, Calcium Release-Activated Calcium Modulator 1; siRNA, small interfering RNA; STIM1, Stromal interacting molecule 1; Th2, T helper 2 cells; WT, Wild-type.
Ethics Approval and Consent to Participate
This manuscript contains samples collected from human subjects that have been approved by the Medical Ethics Committee of Zhongnan Hospital (approval number: 2019044). All procedures performed in this study involving human participants were in accordance with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. The animal experiments were approved by Animal Ethics Committee of Zhongnan Hospital (Approval number: 20200708). The ethics committee reviewed the research protocol based on Institutional Animal Care and Use Committee guidelines.
Acknowledgments
We thank Dr. Xue-feng Zhou from Department of Thoracic Surgery of Zhongnan Hospital, Wuhan University for his help in samples collection. We thank Leonie McKinlay for the English editing.
Author Contributions
We have previously demonstrated that type 2 inflammatory cytokine IL-13 regulates airway remodeling in asthma by acting on store-operated Ca2+ entry (SOCE). Oria1 is the pore-forming subunit of SOC channels. The bioinformatic analysis identified not only an interaction between miR-93-5p and LncRNA H19 but also an interaction between miR-93-5p and Orai1. In cultured human bronchial smooth muscle cells (HBSMCs), we found that IL-13 concentration-dependently increased the expression of H19 and Orai1 and decreased the expression of miR-93-5p. H19 knockdown with siRNA counteracted the promoting effects of IL-13 on the expression of miR-93-5p and Orai1, and proliferation and migration of hBSMCs. Moreover, the effect of IL-13 on the expression of H19 and Orai1 was attenuated by miR-93-5p mimic and increased by miR-93-5p inhibitor. IL-13-promoted proliferation of hBSMCs was increased by miR-93-5p inhibitor but not affected by miR-93-5p mimic; whereas IL-13-promoted migration of hBSMCs was increased by miR-93-5p inhibitor and attenuated by miR-93-5p mimic. The inhibiting effect of H19 knockdown on IL-13-induced Orai1 expression, the proliferation and migration of hBSMCs was counteracted by miR-93-5p inhibitor but not impacted by miR-93-5p mimic. In asthma mice models, we found that the expression of H19 and Orai1 was higher in the lungs of asthma mice than in control mice. In acute asthma mice, H19 siRNA reduced Orai1 expression, inflammatory cell infiltration and goblet cell hyperplasia in the lungs. In chronic asthma mice, H19 siRNA reduced Orai1 expression, inflammatory cell infiltration, goblet cell hyperplasia, collagen deposition and smooth muscle mass in the lungs. We provide evidence to demonstrate a novel regulating mechanism for the activity of SCOE in ASMCs and propose that H19 may represent a potential therapeutic target for airway remodeling in asthma by regulating SOCE.
Funding
This study was supported by Zhongnan Hospital of Wuhan University Science, Technology and Innovation Seed Fund, Project number: znpy2018110.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Papi A, Brightling C, Pedersen SE, Reddel HK. Asthma. Lancet. 2018;391(10122):783–800.
2. Akdis CA, Arkwright PD, Bruggen MC, et al. Type 2 immunity in the skin and lungs. Allergy. 2020;75(7):1582–1605.
3. Saglani S, Lui S, Ullmann N, et al. IL-33 promotes airway remodeling in pediatric patients with severe steroid-resistant asthma. J Allergy Clin Immunol. 2013;132(3):676–685 e613.
4. Manson ML, Safholm J, James A, et al. IL-13 and IL-4, but not IL-5 nor IL-17A, induce hyperresponsiveness in isolated human small airways. J Allergy Clin Immunol. 2020;145(3):808–817 e802.
5. Wang X, Xu C, Ji J, et al. IL-4/IL-13 upregulates Sonic hedgehog expression to induce allergic airway epithelial remodeling. Am J Physiol Lung Cell Mol Physiol. 2020;318(5):L888–L899.
6. Doucet C, Brouty-Boye D, Pottin-Clemenceau C, Canonica GW, Jasmin C, Azzarone B. Interleukin (IL) 4 and IL-13 act on human lung fibroblasts. Implication in asthma. J Clin Invest. 1998;101(10):2129–2139.
7. McKnight CG, Potter C, Finkelman FD. IL-4Ralpha expression by airway epithelium and smooth muscle accounts for nearly all airway hyperresponsiveness in murine allergic airway disease. Mucosal Immunol. 2020;13(2):283–292.
8. Wang YM, Xu WJ, Xiang LL, et al. Store-operated calcium entry-associated regulatory factor regulates airway inflammation and airway remodeling in asthma mice models. Am J Physiol Lung Cell Mol Physiol. 2021;321(3):L533–L544.
9. Gao YD, Zou JJ, Zheng JW, et al. Promoting effects of IL-13 on Ca2+ release and store-operated Ca2+ entry in airway smooth muscle cells. Pulm Pharmacol Ther. 2010;23(3):182–189.
10. Prakriya M, Lewis RS. Store-Operated Calcium Channels. Physiol Rev. 2015;95(4):1383–1436.
11. Zou JJ, Gao YD, Geng S, Yang J. Role of STIM1/Orai1-mediated store-operated Ca(2)(+) entry in airway smooth muscle cell proliferation. J Appl Physiol. 2011;110(5):1256–1263.
12. Spinelli AM, Gonzalez-Cobos JC, Zhang X, et al. Airway smooth muscle STIM1 and Orai1 are upregulated in asthmatic mice and mediate PDGF-activated SOCE, CRAC currents, proliferation, and migration. Pflugers Arch. 2012;464(5):481–492.
13. Sathish V, Abcejo AJ, Thompson MA, Sieck GC, Prakash YS, Pabelick CM. Caveolin-1 regulation of store-operated Ca(2+) influx in human airway smooth muscle. Eur Respir J. 2012;40(2):470–478.
14. Atianand MK, Caffrey DR, Fitzgerald KA. Immunobiology of Long Noncoding RNAs. Annu Rev Immunol. 2017;35:177–198.
15. Ali T, Grote P. Beyond the RNA-dependent function of LncRNA genes. Elife. 2020;9:548.
16. Zhu X, Wei Y, Dong J. Long Noncoding RNAs in the Regulation of Asthma: current Research and Clinical Implications. Front Pharmacol. 2020;11:532849.
17. Zheng M, Hong W, Gao M, et al. Long Noncoding RNA COPDA1 Promotes Airway Smooth Muscle Cell Proliferation in Chronic Obstructive Pulmonary Disease. Am J Respir Cell Mol Biol. 2019;61(5):584–596.
18. Mao Z, Shi Y, Cao Q, et al. Transcriptional regulation on the gene expression signature in combined allergic rhinitis and asthma syndrome. Epigenomics. 2018;10(2):119–131.
19. Su H, Xu X, Yan C, et al. LncRNA H19 promotes the proliferation of pulmonary artery smooth muscle cells through AT1R via sponging let-7b in monocrotaline-induced pulmonary arterial hypertension. Respir Res. 2018;19(1):254.
20. Lopez-Urrutia E, Bustamante Montes LP. Ladron de Guevara Cervantes D, Perez-Plasencia C, Campos-Parra AD. Crosstalk Between Long Non-coding RNAs, Micro-RNAs and mRNAs: deciphering Molecular Mechanisms of Master Regulators in Cancer. Front Oncol. 2019;9:669.
21. Ma J, Zhang L, Hao J, Li N, Tang J, Hao L. Up-regulation of microRNA-93 inhibits TGF-beta1-induced EMT and renal fibrogenesis by down-regulation of Orai1. J Pharmacol Sci. 2018;136(4):218–227.
22. Lu Q, Guo Z, Xie W, et al. The lncRNA H19 Mediates Pulmonary Fibrosis by Regulating the miR-196a/COL1A1 Axis. Inflammation. 2018;41(3):896–903.
23. Xiao Y, Liu R, Li X, et al. Long Noncoding RNA H19 Contributes to Cholangiocyte Proliferation and Cholestatic Liver Fibrosis in Biliary Atresia. Hepatology. 2019;70(5):1658–1673.
24. Woo LN, Guo WY, Wang X, et al. A 4-Week Model of House Dust Mite (HDM) Induced Allergic Airways Inflammation with Airway Remodeling. Sci Rep. 2018;8(1):6925.
25. Wills-Karp M, Luyimbazi J, Xu X, et al. Interleukin-13: central mediator of allergic asthma. Science. 1998;282(5397):2258–2261.
26. Ingram JL, Kraft M. IL-13 in asthma and allergic disease: asthma phenotypes and targeted therapies. J Allergy Clin Immunol. 2012;130(4):829–842.
27. Sugita K, Steer CA, Martinez-Gonzalez I, et al. Type 2 innate lymphoid cells disrupt bronchial epithelial barrier integrity by targeting tight junctions through IL-13 in asthmatic patients. J Allergy Clin Immunol. 2018;141(1):300–310 e311.
28. Yang SJ, Allahverdian S, Saunders ADR, Liu E, Dorscheid DR. IL-13 signaling through IL-13 receptor alpha2 mediates airway epithelial wound repair. FASEB J. 2019;33(3):3746–3757.
29. Sherrill JD, Kiran KC, Blanchard C, et al. Analysis and expansion of the eosinophilic esophagitis transcriptome by RNA sequencing. Genes Immun. 2014;15(6):361–369.
30. Roy S, Schmeier S, Kaczkowski B, et al. Transcriptional landscape of Mycobacterium tuberculosis infection in macrophages. Sci Rep. 2018;8(1):6758.
31. Yue L, Yin X, Hao F, et al. Long Noncoding RNA Linc00632 Inhibits Interleukin-13-Induced Inflammatory Cytokine and Mucus Production in Nasal Epithelial Cells. J Innate Immun. 2020;12(1):116–128.
32. Wang L, Lv Q, Song X, Jiang K, Zhang J. ADRB2 suppresses IL-13-induced allergic rhinitis inflammatory cytokine regulated by miR-15a-5p. Hum Cell. 2019;32(3):306–315.
33. Su Y, Wang J, Zou J, Han W, Li S. miR-330 regulates interleukin-13-induced MUC5AC secretion by targeting Munc18b in human bronchial epithelial cells. Int J Clin Exp Pathol. 2018;11(7):3463–3470.
34. Solberg OD, Ostrin EJ, Love MI, et al. Airway epithelial miRNA expression is altered in asthma. Am J Respir Crit Care Med. 2012;186(10):965–974.
35. Chiba Y, Tanabe M, Goto K, Sakai H, Misawa M. Down-regulation of miR-133a contributes to up-regulation of Rhoa in bronchial smooth muscle cells. Am J Respir Crit Care Med. 2009;180(8):713–719.
36. Chiba Y, Ando Y, Kato Y, Hanazaki M, Sakai H. Down-regulation of miR-140-3p is a cause of the interlukin-13-induced up-regulation of RhoA protein in bronchial smooth muscle cells. Small GTPases. 2021;1:1–6.
37. Yu H, Qi N, Zhou Q. LncRNA H19 Inhibits Proliferation and Migration of Airway Smooth Muscle Cells Induced by PDGF-BB Through miR-21/PTEN/Akt Axis. J Asthma Allergy. 2021;14:71–80.
38. Bousquet J, Agache I, Blain H, et al. Management of anaphylaxis due to COVID-19 vaccines in the elderly. Allergy. 2021;1:55.
39. Dey BK, Pfeifer K, The DA. H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28(5):491–501.
40. Abdelmohsen K, Srikantan S, Tominaga K, et al. Growth inhibition by miR-519 via multiple p21-inducing pathways. Mol Cell Biol. 2012;32(13):2530–2548.
41. Deng W, Wang J, Zhang J, Cai J, Bai Z, Zhang Z. Orai1, a Direct Target of microRNA-519, Promotes Progression of Colorectal Cancer via Akt/GSK3beta Signaling Pathway. Dig Dis Sci. 2016;61(6):1553–1560.
42. Zhang S, Al-Maghout T, Cao H, et al. Gut Bacterial Metabolite Urolithin A (UA) Mitigates Ca(2+) Entry in T Cells by Regulating miR-10a-5p. Front Immunol. 2019;10:1737.
43. Zhang S, Al-Maghout T, Zhou Y, et al. Role of Dicer Enzyme in the Regulation of Store Operated Calcium Entry (SOCE) in CD4+ T Cells. Cell Physiol Biochem. 2016;39(4):1360–1368.
44. Miao C, Bai L, Yang Y, Huang J. Dysregulation of lncRNAs in Rheumatoid Arthritis: biomarkers, Pathogenesis and Potential Therapeutic Targets. Front Pharmacol. 2021;12:652751.
45. Zhang X, Ji S, Cai G, et al. H19 Increases IL-17A/IL-23 Releases via Regulating VDR by Interacting with miR675-5p/miR22-5p in Ankylosing Spondylitis. Mol Ther Nucleic Acids. 2020;19:393–404.
46. Wang B, Suen CW, Ma H, et al. The Roles of H19 in Regulating Inflammation and Aging. Front Immunol. 2020;11:579687.
47. Gamaev L, Mizrahi L, Friehmann T, et al. The pro-oncogenic effect of the lncRNA H19 in the development of chronic inflammation-mediated hepatocellular carcinoma. Oncogene. 2021;40(1):127–139.
48. Lan X, Sun W, Dong W, et al. Downregulation of long noncoding RNA H19 contributes to the proliferation and migration of papillary thyroid carcinoma. Gene. 2018;646:98–105.
49. Chen C, Liu WR, Zhang B, et al. LncRNA H19 downregulation confers erlotinib resistance through upregulation of PKM2 and phosphorylation of AKT in EGFR-mutant lung cancers. Cancer Lett. 2020;486:58–70.
50. Yuan Y, Li X, Chu Y, Ye G, Yang L, Dong Z. Long Non-coding RNA H19 Augments Hypoxia/Reoxygenation-Induced Renal Tubular Epithelial Cell Apoptosis and Injury by the miR-130a/BCL2L11 Pathway. Front Physiol. 2021;12:632398.
51. Austin PJ, Tsitsiou E, Boardman C, et al. Transcriptional profiling identifies the long noncoding RNA plasmacytoma variant translocation (PVT1) as a novel regulator of the asthmatic phenotype in human airway smooth muscle. J Allergy Clin Immunol. 2017;139(3):780–789.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.