Back to Journals » OncoTargets and Therapy » Volume 9
IL-10-1082A/G, -592C/A, and -819T/C polymorphisms in association with lung cancer susceptibility: a meta-analysis
Received 29 July 2016
Accepted for publication 29 August 2016
Published 7 October 2016 Volume 2016:9 Pages 6083—6091
DOI https://doi.org/10.2147/OTT.S118481
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Min Li
Liang Liu,1 Feng Zheng2
1Department of Thoracic Surgery, The TCM Hospital of Changzhou Affiliated to Nanjing University of TCM, Changzhou, Jiangsu, People’s Republic of China; 2Department of Critical Care Medicine, The Third Affiliated Hospital of Soochow University, The First People’s Hospital of Changzhou, Changzhou, Jiangsu, People’s Republic of China
Abstract: Numerous studies have examined the association between interleukin-10 (IL-10 -1082A/G, -592C/A, and -819T/C) gene polymorphisms and risk of lung cancer, but these have revealed inconsistent results. The aim of this study was to clarify the relationship between these polymorphisms and the risk of lung cancer by performing a meta-analysis. The published literature concerning IL-10 polymorphisms and lung cancer risk were retrieved by systematically searching PubMed, Embase, Web of Science, China National Knowledge Infrastructure, WanFang, and Database of Chinese Scientific and Technical Periodicals (VIP) database. Statistical analysis was conducted with Stata 12.0 software. A total of ten published articles comprising of 19 studies were selected, including seven studies (1,960 controls and 1,321 cases) for IL-10 -1082A/G, seven studies (2,613 controls and 1,839 cases) for IL-10 -592C/A, and five studies (1,558 controls and 926 cases) for IL-10 -819T/C. This study found that the IL-10 -1082A/G and -592C/A polymorphisms were significantly associated with the risk of lung cancer in the overall analysis. When stratified by ethnicity, significantly increased risks were observed for IL-10 -1082A/G, -592C/A, and -819T/C polymorphisms in Asians (for -1082A/G, AA vs [AG + GG]: odds ratio [OR] =1.20, confidence interval [CI] =1.05–1.39, P<0.05; for C-592A, C vs A, OR =1.36, CI =1.20–1.53, P<0.05; CC vs AA, OR =1.85, CI =1.45–2.37, P<0.05; CC vs [CA + AA], OR =1.36, CI =1.15–1.61, P<0.05; for -819T/C, T vs C: OR =1.21, CI =1.06–1.38, P<0.05; TT vs CC, OR =1.54, CI =1.18–2.01, P<0.05; [TT + TC] vs CC, OR =1.51, CI =1.17–1.95, P<0.05). Moreover, the data indicated that there was a significant association between IL-10 -819T/C polymorphism and non-small-cell lung cancer risk. No significant publication bias was detected under the four genetic models (allele model, homozygous model, dominant model, and recessive model) in this meta-analysis. On the basis of these 19 studies, this study found that the IL-10 -1082A/G and -819T/C polymorphisms might have a significant association with risk of lung cancer in Asian populations.
Keywords: interleukin-10, lung neoplasms, polymorphism, susceptibility, meta-analysis
Introduction
Lung cancer is one of the most commonly diagnosed tumors and continually leads to many deaths worldwide.1 There were an estimated 1.8 million new cases of lung cancer in 2012, which occurred in the developing countries, accounting for 58% of the total cases. An estimated 1,987,909 new cases of lung cancer were diagnosed and 1,732,185 deaths were due to lung cancer in 2012. Moreover, lung cancer is the most common tumor in males.2 The incidence of lung cancer is still obviously increasing.3 The 5-year survival rate for lung cancer patients was only 16% in the past four decades, and the main obstacle in improving lung cancer prognosis was delayed diagnosis.4 There are many reasons for the occurrence of lung cancer, but the major external risk factor was easily identified – cigarette smoking. It has been reported that current smokers are approximately 20 times more likely than never smokers to develop lung cancer.5,6 Thus, smoking cessation is very important for public health. In addition, early detection and diagnosis play a crucial role in decreasing the mortality of lung cancer. Survival of lung cancer patients undergoing lung resection was more than 80%, which suggested that early detection and diagnosis helped immensely in the early treatment of lung cancer.7,8 Increasing evidence has indicated that polymorphisms of numerous susceptible genes play a significant association in the development of lung cancer. Thus, these polymorphisms could be useful as biomarkers for lung cancer detection.
Interleukin-10 (IL-10) cytokine, which is essential for TH2 responses, plays a central role in anti-inflammation and is also capable of blocking tumor immune surveillance to inhibit T-cell immunity.9 A recent study has shown that the expression of IL-10 was associated with progression of tumors including lung cancer.10 Some genetic polymorphisms of IL-10 gene, especially in the promoter region, could affect the expression of gene-encoded proteins associated with lung cancer susceptibility and prognosis.11 Three SNPs at positions -1082, -592, and -819 of IL-10 gene promoter have been reported to be associated with expression of IL-10 gene.12 Moreover, many individual studies have investigated the role of IL-10 -1082A/G, -592C/A, and -819T/C polymorphisms in lung cancer risk. Nevertheless, the results of these studies remained ambiguous and inconclusive owing to the reduced power of single studies. In this study, an updated meta-analysis was performed to estimate the effect of IL-10 -1082A/G, -592C/A, and -819T/C polymorphisms on lung cancer risk.
Material and methods
Literature search strategy
The databases of PubMed, Embase, Web of Science, China National Knowledge Infrastructure, WanFang, and Database of Chinese Scientific and Technical Periodicals (VIP) were searched up to July 2016 to identify relevant studies published concerning the correlation of IL-10 -1082A/G, -592 C/A, and -819T/C polymorphisms with lung cancer risk. The following MESH terms and their synonyms were used: (“Lung Neoplasms” [MESH] or “lung cancer” or “lung tumor” or “lung carcinoma”) and (“Interleukin-10” or “IL-10”). Language restriction was not applied in the process of retrieval. The reference lists of retrieved article were hand searched.
Inclusion criteria and exclusion criteria
The following inclusion criteria were used: 1) studies investigating the association between IL-10 -1082A/G, -592C/A, and -819T/C polymorphisms and lung cancer risk; 2) studies with a case–control or cohort design; 3) studies with sufficient genotyping data available for calculation of odds ratios (ORs) and 95% confidence intervals (95% CIs); and 4) studies performed in humans. The exclusion criteria were as follows: 1) not a case–control or cohort study, 2) studies without available genotype data, 3) studies with duplicate data, and 4) meta-analysis and reviews.
Quality score assessment
The Newcastle–Ottawa scale was used to assess the quality of studies included. Two investigators independently calculated the score of each study. The scores ranged from 0 to 9, and a score of ≥7 indicated a high quality study.
Data extraction
Two investigators independently searched articles and extracted the original data according to the inclusion criteria and exclusion criteria. The extracted information from each eligible study included the following: 1) the first author’s name, 2) year of publication, 3) ethnicity, 4) source of controls, 5) number of cases and controls, and 6) genotype frequencies. Disagreement in data extraction was settled by consulting with the third investigator.
Statistical analysis
The strength of association between IL-10 -1082A/G, -592C/A, and -819T/C polymorphisms and lung cancer risk was evaluated by calculating crude ORs and 95% CIs under an allele, a homozygous, a dominant, and a recessive model, respectively. Chi-square-based Q-test and I2 statistic test were performed to evaluate the heterogeneity assumption. A random-effects model (the DerSimonian and Laird method) was used to calculate the crude ORs of each study in case of P<0.05 or I2>50%, whereas a fixed-effects model (the Mantel–Haenszel method) was used if heterogeneity did not exist.13,14 Potential publication bias was evaluated using Begg’s funnel plots and Egger’s test. Sensitivity analysis was conducted to estimate the stability of the results by omitting each single study. The overall analysis and stratified analysis were performed using Stata 12.0 software (Stata Corp LP, College Station, TX, USA).
Results
Characteristics of published studies
Based on the inclusion criteria, a total of 372 publications were obtained on literature searching. After removing the articles that did not deal with the association of IL-10 polymorphism with lung cancer risk, reviews, and meta-analysis, 14 were left for further analysis. After further evaluation by reading full texts, four publications were excluded because of insufficient data on IL-10 genotype. Finally, 10 articles with 19 studies that investigated the correlation of IL-10 -1082A/G, -592C/A, and -819T/C polymorphisms with lung cancer risk were used for the further statistical analysis. Of these, eleven studies were conducted in Asians and eight studies were performed in Caucasians. In addition, eight publications were in English and two publications were in Chinese. The characteristics of all the studies included are listed in Table 1 and shown in Figure 1.
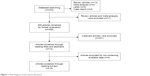  | Figure 1 Flow diagram of the search literature. |
Quantitative analysis
A total of 10 publications amounting to 19 case–control studies were included in this meta-analysis. In the overall analysis, the presence of IL-10 -1082A/G polymorphisms was associated with increased lung cancer risk, while IL-10 -592C/A polymorphisms were shown to decrease the lung cancer risk (for A-1082G, A vs G, OR =1.17, CI =1.04–1.30, P<0.05; AA vs [AG + GG], OR =1.21, CI =1.05–1.40, P<0.05; for C-592A, C vs A, OR =0.84, CI =0.77–0.91, P<0.05; CC vs AA, OR =0.74, CI =0.64–087, P<0.05; CC vs [CA + AA], OR =0.83, CI =0.74–0.94, P<0.05; [CC + CA] vs AA, OR =0.79, CI =0.69–0.91, P<0.05). When stratified by ethnicity, IL-10 -1082A/G, C-592A, and T-819C polymorphisms were associated with increased lung cancer risk in Asians (for -1082A/G, AA vs [AG + GG]: OR =1.20, CI =1.05–1.39, P<0.05; for C-592A, C vs A, OR =1.36, CI =1.20–1.53, P<0.05; CC vs AA, OR =1.85, CI =1.45–2.37, P<0.05; CC vs [CA + AA], OR =1.36, CI =1.15–1.61, P<0.05; for -819T/C, T vs C: OR =1.21, CI =1.06–1.38, P<0.05; TT vs CC, OR =1.54, CI =1.18–2.01, P<0.05; [TT + TC] vs CC, OR =1.51, CI =1.17–1.95, P<0.05). Furthermore, when stratified based on disease type, a significant correlation was detected between IL-10 T-819C polymorphism and non-small-cell lung cancer (NSCLC; for -819T/C, T vs C: OR =1.57, CI =1.15–2.16, P<0.05; TT vs CC, OR =3.15, CI =1.58–6.30, P<0.05; [TT + TC] vs CC, OR =3.07, CI =1.58–5.93, P<0.05). Interestingly, no significant association between IL-10 -1082A/G, C-592A, and T-819C polymorphisms and lung cancer risk was found in Caucasians. Furthermore, significantly decreased risk of lung cancer risk was found in the overall analysis of C-592A polymorphism, whereas significantly increased risk was detected in the Asian population (Table 2).
Heterogeneity analysis
Under the homozygous genetic models, significant heterogeneity existed among the studies on IL-10 -1082A/G and -819T/C polymorphisms (for 1082A/G, I2=71.8%, P=0.007; for -819T/C, I2=88.0%, P=0.000). The forest plots suggested that ethnicity and disease type might be one of the sources of heterogeneity for IL-10 -1082A/G and -819T/C polymorphisms. However, heterogeneity was significantly lowered after the subgroup analyses based on ethnicity and disease type were conducted. This indicated that the ethnicity and disease type might lead to the heterogeneity. Furthermore, a meta-regression revealed that score, sample size, and genotyping method were not the sources of heterogeneity (for IL-10 -1082A/G and -819T/C, P>0.05; Figures 2–4).
  | Figure 2 Forest plot of lung cancer risk associated with IL-10 -1082A/G (AA vs GG) polymorphism stratified by ethnicity. |
  | Figure 3 Forest plot of lung cancer risk associated with IL-10 C-592A (CC vs AA) polymorphism stratified by ethnicity. |
  | Figure 4 Forest plot of lung cancer risk associated with IL-10 -819T/C (TT vs CC) polymorphism stratified by ethnicity. |
Publication bias and sensitivity analysis
On the basis of the Begg’s test and Egger’s test, no publication bias was found for the association between IL-10 -1082A/G, -592C/A, and -819T/C polymorphisms and lung cancer risk. Moreover, sensitivity analysis indicated that the results were stable after sequentially omitting each study (Figures 5–7).
  | Figure 5 Begg’s funnel plot with pseudo 95% confidence limits for studies of the association between lung cancer risk and IL-10 -1082A/G (AA vs GG) polymorphism. |
  | Figure 6 Begg’s funnel plot with pseudo 95% confidence limits for studies of the association between lung cancer risk and IL-10 C-592A (CC vs AA) polymorphism. |
  | Figure 7 Begg’s funnel plot with pseudo 95% confidence limits for studies of the association between lung cancer risk and IL-10 -819T/C (TT vs CC) polymorphism. |
Discussion
IL-10 was shown to have a significant effect on the tumorigenesis of different cancers and could even protect tumors by inhibiting the function of macrophages and dendritic cells, which present tumor cell antigens to the cytotoxic T lymphocytes.15 Previous studies have found an association between IL-10 polymorphism and oral cancer, breast cancer, gastric cancer, colorectal cancer, esophageal cancer, hepatocellular cancer, cervical cancer, thyroid cancer, and nasopharyngeal cancer.16–24 As for lung cancer, many studies have revealed that the loss of IL-10 expression could promote progression and poor clinical outcomes of lung cancer; however, the opposite effects were also found in some patients.25 Interestingly, the absence of IL-10 had opposite effects on the early and late stages of NSCLC.26 Hence, the expression or polymorphisms of IL-10 gene might serve as a good indicator of susceptibility and prognostic outcome for lung cancer in different stages of the disease.27 These lung cancer biomarkers might be used for screening, early detection, early diagnosis, prognosis, prediction, stratification, and therapy response monitoring.3 Although previous case–control studies showed that the IL-10 polymorphism might contribute to the risk of lung cancer, and the sample size of the studies was too small. Thus, this meta-analysis was performed.
Through this comprehensive meta-analysis, the association between IL-10 -1082A/G, -592C/A, and -819T/C polymorphism and the risk of lung cancer was evaluated. To our knowledge, this is the first systematic meta-analysis to date that deals with the association between IL-10 -1082A/G, -592C/A, and -819T/C polymorphisms and lung cancer risk. In the overall analysis, we found that IL-10 -1082A/G, -592C/A, and -819T/C polymorphisms were significantly associated with lung cancer risk. Interestingly, IL-10 -1082A/G, -592C/A, and -819T/C polymorphisms were significantly associated with lung cancer risk in Asians, but not in the Caucasians. Furthermore, the results indicated that individuals who carried the IL-10 -1082G allele had an increased risk of lung cancer, and those with the IL-10 -819T allele were shown to have a decreased risk of lung cancer compared to those with the -819C allele. The results of previous case–control studies by Shih et al,28 Seifart et al,29 Colakogullari et al,30 and Peddireddy et al31 for IL-10 -1082A/G polymorphism were consistent with this meta-analysis and indicated that the IL-10 -1082A/G polymorphism could increase the risk of lung cancer. The results of Hsia et al,32 Shih et al,28 and Zhang et al33 for IL-10 -819T/C polymorphism showed that IL-10 -819T/C polymorphism was significantly associated with an increased risk of lung cancer. Nevertheless, different results were also found in the studies of Hsia et al32 and Hart et al34 concerning the IL-10 -1082A/G polymorphism. However, several of the included studies showed no significant correlation of IL-10 A-592C and -819T/C polymorphisms with lung cancer risk.29,30,32,34–36 In addition, there was adequate evidence indicating the different frequency distribution of IL-10 -592C/A and -819T/C allele between Asians and Caucasians in the meta-analysis. And this result was consistent with the data given in the HapMap database. Thus, a decreased risk of lung cancer in Caucasians and an increased risk of lung cancer in Asians was associated with the IL-10 -592C/A and -819T/C polymorphisms, although no significant association of IL-10 -592C/A and -819T/C polymorphisms with lung cancer risk in Caucasians was found. In addition, based on the results of the subgroup analysis, we found that IL-10 -819T/C polymorphism was associated with susceptibility to NSCLC. A previous study reported that IL-10 might be involved in the process of tumor escape from the immune response, and increased production of IL-10 has been shown to correlate with the survival of NSCLC patients.28 However, only one study was included in the subgroup analysis, and this lowered the statistical power. For the IL-10 -1082A/G and -592C/A polymorphism, no significant correlation with NSCLC risk was detected. The analysis of small-cell lung cancer was not performed because the sample of small-cell lung cancer was too small and the classification of small-cell lung cancer was not clear. On the basis of the meta-analysis, to a large extent, there was no significant association between IL-10 -1082A/G, -592C/A polymorphism and NSCLC risk. To confirm this conclusion, studies with larger sample number must be conducted. The study results showed that subgroup analysis is very important for a systematic review and meta-analysis. From the studies included, there is evidence that the association between IL-10 polymorphism and lung cancer risk is still not clear. Therefore, performing a meta-analysis and systematic review was necessary to clarify this relevance. These results are shown in Figure 3.
Limitations
Several limitations of this meta-analysis should be acknowledged. First, the small number of eligible studies might limit the meta-analysis. There were only five or seven studies available for three SNPs of IL-10 gene in the stratification analysis in particular, which might have attenuated the statistical power of the meta-analysis. Second, the missing individual data, for example, cancer stage and cancer subtype might also have an impact on the interpretation. Third, this meta-analysis could not address the gene–environmental interactions in the correlation of IL-10 polymorphism with lung cancer risk. The conclusions only depend on the ORs, which might lead to confounding bias. Finally, the inclusion of data that was inconsistent with HWE might affect the accuracy of the results. However, the large sample size could weaken the impact of the association of IL-10 polymorphism with lung cancer risk.
In conclusion, this meta-analysis of ten case–control studies suggested that the IL-10 -1082A/G, -592C/A, and -819T/C polymorphisms were associated with the increased risk of lung cancer in Asians. Nevertheless, considering the limitations of the study, further large-scale, well-designed, and population-based studies are still needed to assess the correlation between IL-10 polymorphism and lung cancer risk.
Acknowledgment
The authors would like to thank all the participants involved in the research.
Disclosure
The authors report no conflicts of interest in this work.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. | ||
IAFROC, WHO, GLOBOCAN 2012. Estimated cancer incidence, mortality and prevalence worldwide in 2012: liver cancer. | ||
Chen W, Zhang S, Zou X. Evaluation on the incidence, mortality and tendency of lung cancer in China. Thoracic Cancer. 2010;1:35–40. | ||
I H, Cho JY. Lung cancer biomarkers. Adv Clin Chem. 2015;72:107–170. | ||
Khuder SA. Effect of cigarette smoking on major histological types of lung cancer: a meta-analysis. Lung Cancer. 2001;31:139–148. | ||
Kowgier M, Qiu LA, Tse H. Surgery for small cell lung cancer. Asian Pac J Surg Oncol. 2015;1:171–180. | ||
Wang S, Lan X, Tan S, Wang S, Li Y. P53 codon 72 Arg/Pro polymorphism and lung cancer risk in Asians: an updated meta-analysis. Tumour Biol. 2013;34:2511–2520. | ||
Sandri A, Brunelli A, Rzyman W, et al. Pathology of non-small cell lung cancer and small cell lung cancer. Asian Pac J Surg Oncol. 2015;1:113–124. | ||
Brooks DG, Trifilo MJ, Edelmann KH, Teyton L, McGavern DB, Oldstone MB. Interleukin-10 determines viral clearance or persistence in vivo. Nat Med. 2006;12:1301–1309. | ||
Kim R, Emi M, Tanabe K, Arihiro K. Tumor-driven evolution of immunosuppressive networks during malignant progression. Cancer Res. 2006;66:5527–5536. | ||
Rausch SM, Gonzalez BD, Clark MM, et al. SNPs in PTGS2 and LTA predict pain and quality of life in long term lung cancer survivors. Lung Cancer. 2012;77:217–223. | ||
Stearns ME, Rhim J, Wang M. Interleukin 10 (IL-10) inhibition of primary human prostate cell-induced angiogenesis: IL-10 stimulation of tissue inhibitor of metalloproteinase-1 and inhibition of matrix metalloproteinase (MMP)-2/MMP-9 secretion. Clin Cancer Res. 1999;5:189–196. | ||
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. | ||
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. | ||
Goncalves AS, Arantes DA, Bernardes VF, et al. Immunosuppressive mediators of oral squamous cell carcinoma in tumour samples and saliva. Hum Immunol. 2015;76:52–58. | ||
Hsu HJ, Yang YH, Shieh TY, et al. Role of cytokine gene (interferon-gamma, transforming growth factor-beta1, tumor necrosis factor-alpha, interleukin-6, and interleukin-10) polymorphisms in the risk of oral precancerous lesions in Taiwanese. Kaohsiung J Med Sci. 2014;30:551–558. | ||
Tsai CW, Tsai MH, Shih LC, Chang WS, Lin CC, Bau DT. Association of interleukin-10 (IL10) promoter genotypes with nasopharyngeal carcinoma risk in Taiwan. Anticancer Res. 2013;33:3391–3396. | ||
Wang Z, Liu QL, Sun W, et al. Genetic polymorphisms in inflammatory response genes and their associations with breast cancer risk. Croat Med J. 2014;55:638–646. | ||
Yu T, Lu Q, Ou XL, Cao DZ, Yu Q. Clinical study on gastric cancer susceptibility genes IL-10-1082 and TNF-alpha. Genet Mol Res. 2014;13:10909–10912. | ||
Miteva LD, Stanilov NS, Deliysky TS, Stanilova SA. Significance of -1082A/G polymorphism of IL10 gene for progression of colorectal cancer and IL-10 expression. Tumour Biol. 2014;35:12655–12664. | ||
Sakamoto K, Oka M, Yoshino S, et al. Relationship between cytokine gene polymorphisms and risk of postoperative pneumonia with esophageal cancer. J Gastrointest Surg. 2014;18:1247–1253. | ||
Pan D, Zeng X, Yu H, et al. Role of cytokine gene polymorphisms on prognosis in hepatocellular carcinoma after radical surgery resection. Gene. 2014;544:32–40. | ||
Zhang X, Zhang L, Tian C, Yang L, Wang Z. Genetic variants and risk of cervical cancer: epidemiological evidence, meta-analysis and research review. BJOG. 2014;121:664–674. | ||
Cil E, Kumral A, Kanmaz-Ozer M, et al. Interleukin-10-1082 gene polymorphism is associated with papillary thyroid cancer. Mol Biol Rep. 2014;41:3091–3097. | ||
Wang R, Lu M, Zhang J, et al. Increased IL-10 mRNA expression in tumor-associated macrophage correlated with late stage of lung cancer. J Exp Clin Cancer Res. 2011;30:62. | ||
De Vita F, Orditura M, Galizia G, et al. Serum interleukin-10 levels as a prognostic factor in advanced non-small cell lung cancer patients. Chest. 2000;117:365–373. | ||
Hatanaka H, Abe Y, Kamiya T, et al. Clinical implications of interleukin (IL)-10 induced by non-small-cell lung cancer. Ann oncol. 2000;11:815–819. | ||
Shih CM, Lee YL, Chiou HL, et al. The involvement of genetic polymorphism of IL-10 promoter in non-small cell lung cancer. Lung Cancer. 2005;50:291–297. | ||
Seifart C, Plagens A, Dempfle A, et al. TNF-alpha, TNF-beta, IL-6, and IL-10 polymorphisms in patients with lung cancer. Dis Markers. 2005;21:157–165. | ||
Colakogullari M, Ulukaya E, Yilmaztepe OA, et al. The involvement of IL-10, IL-6, IFN-gamma, TNF-alpha and TGF-beta gene polymorphisms among Turkish lung cancer patients. Cell Biochem Funct. 2011;26:283–290. | ||
Peddireddy V, Badabagni SP, Sulthana S, Kolla VK, Gundimeda SD, Mundluru H. Association of TNFalpha-308, IFNgamma+874, and IL10-1082 gene polymorphisms and the risk of non-small cell lung cancer in the population of the South Indian state of Telangana. Int J Clin Oncol. 2016. Epub ahead of print. | ||
Hsia TC, Chang WS, Liang SJ, et al. Interleukin-10 (IL-10) promoter genotypes are associated with lung cancer risk in Taiwan males and smokers. Anticancer Res. 2014;34:7039–7044. | ||
Zhang YM, Mao YM, Sun YX. Genetic polymorphisms of IL-6 and IL-10 genes correlate with lung cancer in never-smoking Han population in China. Int J Clin Exp Med. 2015;8:1051–1058. | ||
Hart K, Landvik NE, Lind H, Skaug V, Haugen A, Zienolddiny S. A combination of functional polymorphisms in the CASP8, MMP1, IL10 and SEPS1 genes affects risk of non-small cell lung cancer. Lung Cancer. 2011;71:123–129. | ||
Liang HG, Fu SR, Qi XM. Relationship between the genetic polymorphism in Interleukin-10 -592C/A and susceptibility to non-small cell lung cancer. China J Modern Med. 2011;21:189–193. Article in Chinese. | ||
Vogel U, Christensen J, Wallin H, et al. Polymorphisms in genes involved in the inflammatory response and interaction with NSAID use or smoking in relation to lung cancer risk in a prospective study. Mutat Res. 2008;639:89–100. | ||
Hao MG, Wang WJ, Lu Y, et al. [Association of genetic polymorphism within promoter region of TNF-α, IL-1β and IL-10 with the susceptibility to lung cancer]. J Environ Occup Med. 2009;26:24–27. Chinese. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


