Back to Journals » Cancer Management and Research » Volume 10
Identification of WDR12 as a novel oncogene involved in hepatocellular carcinoma propagation
Authors Yin Y , Zhou L, Zhan R, Zhang Q, Li M
Received 4 June 2018
Accepted for publication 10 July 2018
Published 27 September 2018 Volume 2018:10 Pages 3985—3993
DOI https://doi.org/10.2147/CMAR.S176268
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Kenan Onel
Yancun Yin,1 Ling Zhou,2 Renhui Zhan,3 Qiang Zhang,3 Minjing Li3
1Taishan Scholar Immunology Program, School of Basic Medical Sciences, Binzhou Medical University, Yantai 264003, Shandong, China; 2The Key Laboratory of Traditional Chinese Medicine Prescription Effect and Clinical Evaluation of State Administration of Traditional Chinese Medicine, School of Pharmacy, Binzhou Medical University, Yantai 264003, Shandong, China; 3Medicine and Pharmacy Research Center, Binzhou Medical University, Yantai 264003, Shandong, China
Background: Hepatocellular carcinoma (HCC) is one of the most common malignant cancer worldwide. Importantly, the precise mechanisms causing HCC pathogenicity are still unknown. The identification of potential oncogenes plays significant roles in finding novel therapeutic targets for human HCC.
Purpose: WDR12 (WD repeat protein 12), a member of WD repeats family, plays crucial roles in the ribosome biogenesis pathway. However, Whether WDR12 contributes to HCC development remains unknown. The objective of this study was to elucidate the role of WDR12 in HCC development.
Methods: The expression level of WDR12 in HCC tissues and adjacent non-tumor tissues were detected form Gene Expression Omnibus (GEO) database. The expression level of WDR12 in HCC cell lines were examined by RT-PCR and western blot. Kaplan-Meier analysis were used to analyze the effect of WDR12 level on overall and disease-free survival of HCC patients. To examine whether WDR12 supports development of HCC, we inhibited expression of WDR12 by using an shRNA-encoding lentivirus system. Effects of WDR12 knockdown were evaluated on cell-growth, cell-proliferation and cell-migration. The mechanisms involved in HCC cells growth, proliferation and migration were analyzed by western blot assay.
Results: In silico analysis of HCC data sets showed that elevated expression of WDR12 correlated with high serum AFP level, high vascular invasion, high histologic grade and high TNM stage in HCC patients. Furthermore, up-regulated expression of WDR12 significantly correlated with the short overall survival and recurrence time of HCC patients. The shRNA-mediated knockdown of WDR12 expression resulted in reduced proliferation and migration of HepG2 and Huh-7 cells. Notably, inhibition of WDR12 resulted in decreased phosphorylation of AKT, mTOR and S6K1.
Conclusion: Our study indicates that WDR12 contributes to HCC propagation, and indicates that suppression of WDR12 may be a potential strategy for human HCC treatment.
Keywords: hepatocellular carcinoma, WD-repeat proteins, proliferation, migration
Introduction
Hepatocellular carcinoma (HCC), the most common cause of primary liver malignancy, has rapidly increased in recent years.1 In 2012, an estimated 1.5 million new liver cancer cases and deaths occurred worldwide, with China alone accounting for ~50% of the total number.1,2 Furthermore, the incidence of HCC is increasing in Western Europe and North America, which historically have had low rates.2 Although various oncogenes or signaling pathways are implicated in HCC, the molecular basis for HCC initiation and propagation remains largely unknown.2 Therefore, novel therapeutic approaches and molecular targets are urgently required for the effective treatment of human HCC.
Many altered oncogenes or tumor suppressor genes participate in the occurrence and development of human HCC. Mammalian WD-repeat protein 12 (WDR12), a member of the family of proteins ending with tryptophan–aspartic acid (WD) repeats, is a putative homologue of yeast Ytm1p and contains 7 WD repeats. WDR12 mRNA is ubiquitously expressed during embryonic development, and high expression levels are found in the thymus and testis tissue of adult mice.3 In mammals, WDR12 forms a stable complex with Pes1 and Bop1 named PeBoW, which plays crucial roles in the ribosome biogenesis pathway. As a component of PeBoW, WDR12 is an essential factor for the processing of 32S precursor ribosomal RNA and cell proliferation.3–5 Furthermore, WDR12 is required for the formation of 28S and 5.8S ribosomal RNAs and maturation of the 60S ribosomal subunit.5,6 It was observed that knockdown of WDR12 expression in neonatal myocytes resulted in reduced phosphorylation of ERK1/2, p38 MAPK, and HSP27. This finding may partially elucidate the mechanistic role of WDR12 in the regulation of cell proliferation and survival.7,8 Despite these elegant observations, the role of WDR12 in other physiological processes is largely unresolved.9
In the present study, we first found that a high expression level of WDR12 is correlated with negative clinical outcomes in HCC patients. Next, we discovered that WDR12 supports the growth of human HCC cells. Finally, inhibition of WDR12 expression significantly decreased the proliferation of HCC cells by mediating the Akt-mTOR-S6K1 signaling axis. These findings suggest that WDR12 is a promising target for human HCC treatment.
Materials and methods
Cell culture
All the HCC cell lines and the normal hepatocyte LO2 cell line were purchased from Cell Lines Bank, Chinese Academy of Science (Shanghai, China). Cell lines were cultured in DMEM supplemented with 10% FBS at 37°C in 5% CO2. The Ethical Committee of Binzhou Medical University approved the use of the cell lines in this study.
Reagents and antibodies
Anti-human WDR12 antibody (catalog number ab95070) was purchased from Abcam (Cambridge, UK). Antibodies against Akt (catalog number 3063), p-Akt (S473) (catalog number 4060), p-mTOR (S2448) (catalog number 5536), and p-S6K1 (S371) (catalog number 9208) were provided by Cell Signaling Technology (Beverly, MA, USA). Antibodies against S6K1 (catalog number 14485-1-AP), mTOR (catalog number 20657-1-AP), and actin (catalog number 60008-1-Ig) were from ProteinTech Group (Chicago, IL, USA). Secondary antibodies were provided by Zen Bioscience (Chengdu, China).
Gene expression and survival analysis
Microarray data for WDR12 in human HCC tumor or normal liver tissues were obtained from the Gene Expression Omnibus (GEO) data set accession numbers GSE5363, GSE19665, and GSE6764. For survival analyses, HCC data were obtained from the CBioPortal data set “Liver Hepatocellular Carcinoma” (the Cancer Genome Atlas [TCGA]), Provisional (http://www.cbioportal.org/study?id=lihc_tcga), as previously described.10 The most recent update was on June 02, 2016. Primary clinical data from HCC patients were submitted (Supplementary material). Samples with unabridged data were divided into three groups equally according to the mRNA level of WDR12. Kaplan–Meier analysis was performed to estimate the overall and disease-free survival of HCC patients.
Quantitative real-time PCR
Total RNA was extracted using Trizol (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA). Appropriate amounts of RNAs (1 µg) were used to generate cDNA with random hexamers from mRNA (Life Technologies, Thermo Fisher Scientific). The mRNA was quantified using SYBR Green PCR Master Mix (Life Technologies) and normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA levels.
Lentivirus construction and infection
shRNAs targeting WDR12 were inserted into the lentiviral vector PLL3.7. The target sequence of WDR12 was as follows: WDR12-1: 5′-CCTTGGAAGGAAAGTCAATAA-3′ and WDR12-2: 5′-CTTCTATGGATCAGACTATTC-3′. Construction and infection of the lentivirus was performed as previously described.11 The WDR12 shRNA plasmid was mixed with pSPAX2 and pMD2G packaging plasmids at a ratio of 4:3:1 and then transfected into HEK-293T cells using PolyJet reagent. Supernatant containing lentivirus was collected separately at 48 and 72 hours posttransfection. The lentivirus was filtered and used for subsequent infection assays.
Western blot
HCC cells were harvested and lysed in ice-cold RIPA buffer including phosphatase and protease inhibitors (phosSTOP, phenylmethylsulfonyl fluoride [PMSF], and aprotinin). The cell suspension was placed on ice for 20 minutes and then centrifuged for 10 minutes at 14,000 rpm. The supernatant was harvested, and the protein concentration was analyzed using a BCA protein assay kit. The samples were boiled with loading buffer and then separated on SDS-PAGE gels (Bio-Rad Laboratories Inc., Hercules, CA, USA) and transferred to polyvinylidene difluoride (PVDF) membranes. The PVDF membranes were blocked with 5% BSA and then incubated with specific primary antibodies and appropriate secondary antibodies. The blots were detected using chemiluminescent agents with Alpha Innotech’s (San Leandro, CA, USA) FluorChem imaging system. To ensure that equal amounts of sample protein were used for electrophoresis, actin was used as an internal control.
Cell growth/proliferation assay
HCC cells were preinfected with scrambled or WDR12 shRNA, and then cell growth and proliferation were detected. For the cell growth assay, cells were seeded into 24-well plates at 24 hours postinfection. Then, the cell number was counted using a TC20 automated cell counter at the indicated days, and cell growth was evaluated by calculating the fold change of the cell number. Cell proliferation was evaluated using the EdU Kit (catalog number C10310–1; Ribobio, Guangzhou, China) as we previously described.12,13
Transwell assay
Transwell assay was used to determine the migration ability of the HCC cells. HCC cells were preinfected with scrambled or WDR12 shRNA for 24 hours. Then, 3×104 cells were seeded into the top chamber in serum-free DMEM. A total of 750 µL DMEM containing 10% FBS was added into the bottom chambers. After 8 hours of incubation, the cells in the top chamber on the layer were completely removed with Q-tips. The cells beneath the upper chambers were dipped in methanol and stained with 0.25% crystal violet, and then the cell numbers were counted to evaluate the migration ability of the cells.
Statistical analysis
For all experiments, at least three independent repeats were performed, and the data are presented as the mean±SD. Statistical analysis was performed with SPSS 17.0. Student’s t-test was used to compare the differences between two groups, and a one-way ANOVA was used to compare multiple groups. A log-rank test was used to analyze overall and disease-free survival differences between patient groups. A chi-squared test was used to analyze the correlation of WDR12 expression with the clinicopathological features of HCC patients. P-values <0.05 were defined as significantly different.
Results
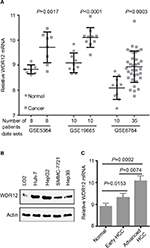
WDR12 is upregulated in HCC samples
To confirm the effect of WDR12 on human HCC patients, we first analyzed the mRNA expression level of WDR12 using an in-silico analysis method. The GEO analysis results indicated that the expression level of WDR12 mRNA in HCC samples was much higher than that in normal tissues (Figure 1A). Next, the level of WDR12 was examined in several HCC cell lines, such as Hep3B, HepG2, Huh-7, and SMMC-7721. We found that WDR12 protein was aberrantly upregulated in human HCC cell lines compared to the expression in normal hepatocyte LO2 (Figure 1B). Then, we further analyzed the mRNA expression level of WDR12 using data from the GEO data set accession number GSE6764. Consistently, WDR12 was more elevated in advanced HCC compared to the levels in early HCC and normal samples (Figure 1C). Taken together, these data show that WDR12 is upregulated in HCC samples.
WDR12 overexpression predicts poor prognosis in HCC patients
To identify whether the elevated levels of WDR12 expression in HCC samples and cell lines correlate with clinical parameters, we analyzed the correlation between the level of WDR12 and the clinicopathological features of HCC patients. Chi-square analysis indicated that high levels of WDR12 were significantly related to the levels of serum AFP (P=0.0218), histologic grade (P=0.0010), vascular invasion (P=0.0013), and TNM stage (P=0.0017) of HCC patients (Table 1, Supplementary material). No other clinicopathological parameter was found to be associated with WDR12 overexpression. Notably, all the positive correlations with WDR12 expression were aggressive clinicopathological features of patients with HCC. To clarify the relationship between the WDR12 level and clinical outcomes in patients with HCC, Kaplan–Meier analysis was used to explain the association of WDR12 mRNA with clinical end points in HCC patients. Concordantly, the high mRNA level of WDR12 was dramatically related to shorter overall survival (Figure 2A, Supplementary material) and reduced disease-free survival (Figure 2B, Supplementary material). These data indicate that WDR12 may be a convincing indicator for poor prognosis in HCC.
  | Figure 2 Expression of WDR12 negatively correlates with clinical outcomes in human HCC patients. Notes: WDR12 mRNA expression data and clinical data from HCC patients were obtained from cBioPortal (http://www.cbioportal.org/study?id=lihc_tcga#summary). HCC samples were divided into three groups equally based on the log2-transformed expression level of WDR12. The effect of WDR12 on the (A) overall and (B) disease-free survival of HCC patients was analyzed using Kaplan–Meier plots. Abbreviations: HCC, hepatocellular carcinoma; WDR, WD-repeat protein. |
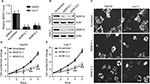
Silencing WDR12 expression inhibits the proliferation of HCC cells
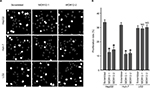
Since WDR12 was most highly expressed in HepG2 and Huh-7 cells among the several assessed cell lines, these two cell lines were used to perform the remaining experiments. To further study the biological role of WDR12, lentiviruses containing shRNAs were used to deplete WDR12 expression. We first determined the efficiency of the designed shRNAs using RT-PCR and Western blot in HepG2 and Huh-7 cells. RT-PCR data indicated that the tested shRNAs efficiently decreased WDR12 transcription compared with scrambled shRNA in both HepG2 and Huh-7 cells (Figure 3A). Concordantly, Western blot results showed similar deletion efficiency at the protein level (Figure 3B). To further explore the effect of WDR12 on HCC, we transfected WDR12 or scrambled shRNAs into HepG2 or Huh-7 cells and then counted the number of cells to measure the cell viability. Treatment with WDR12 shRNA led to a significant decrease in HCC cell growth (Figure 3C). Cells treated with WDR12 shRNAs grew more slowly at 3 days postinfection, and growth inhibition was more apparent at 4 days and 5 days postinfection (Figure 3D and E). For instance, at Days 4 and 5 after infection with shRNAs, the HepG2 cells treated with shRNA1 or shRNA2 had 49.54% and 43.48% or 59.37% and 50.14% growth, respectively, compared with control cells infected with scrambled shRNA (Figure 3D). To investigate the mechanism of how WDR12 promotes the growth of HCC cells, we evaluated cell proliferation and apoptosis in HepG2 and Huh-7 cells infected with scrambled or WDR12 shRNA. 5-Ethynyl-2′-deoxyuridine (EdU) labeling results indicated that downregulation of WDR12 dramatically suppressed the proliferation of HCC cells (Figure 4A and B: 37.33% proliferation cell rate in scrambled shRNA-transfected cells vs 15.47% and 17.36% in WDR12 shRNA1- and shRNA2-treated cells at 72 hours postinfection). However, compared with control cells, WDR12 knockdown had no influence on the apoptosis of HCC cells (data not shown). We also evaluated the biological function of WDR12 in normal liver cells. Interestingly, knockdown of WDR12 had no effect on the proliferation of the normal hepatocyte cell line LO2 (Figure 4A and B). These results indicate that reduced proliferation may significantly slow down the growth of the WDR12-deficient HCC cells.
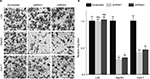
Silencing WDR12 expression inhibits the migration of HCC cells
Since WDR12 expression was markedly associated with TNM and vascular invasion in HCC patients, we hypothesized that WDR12 might affect the motility of HCC cells. Thus, we evaluated the potential role of WDR12 in cell migration using transwell assays. As expected, a significant decrease in transwell cell migration was clearly seen in HepG2 cells transfected with WDR12 shRNA compared to that in control cells transfected with scrambled shRNA (Figure 5A). Statistically, the relative number of migrating cells was significantly decreased in WDR12-deficient cells compared to that in scrambled shRNA-treated cells (Figure 5B). Similar results were observed in another HCC cell line, Huh-7 (Figure 5A and B). However, knockdown of WDR12 had no effect on the migration of a normal liver cell line, LO2 (Figure 5A and B). These results suggest that WDR12 facilitates the migration of HCC cells.
WDR12 promotes Akt-mTOR-S6K1 signaling activation
To identify downstream signaling networks regulated by WDR12, we next examined Akt-mTOR-S6K1 signaling, which plays a significant role in regulating cancer cell proliferation. To inspect the role of WDR12 in the Akt-mTOR-S6K1 signaling pathway, cells were infected with scrambled or WDR12 shRNA, and then the expression and phosphorylation of Akt, mTOR, and S6K1 were analyzed. While knockdown of WDR12 did not induce expression of Akt, mTOR, or S6K1, it decreased the phosphorylation of Akt, mTOR, and S6K1 in both Huh-7 and HepG2 cells (Figure 6A). Finally, HepG2 cells were treated with WDR12 shRNA for 24–72 hours, followed by Western blot analysis of the phosphorylation of Akt, mTOR, and S6K1. We found that knockdown of WDR12 decreased the phosphorylation of Akt, mTOR, and S6K1 in a time-dependent manner (Figure 6B). These findings indicate that WDR12 mediates a signaling network including Akt, mTOR, and S6K1 to promote the growth and proliferation of HCC cells.
Discussion
WD repeats are minimally conserved regions of ~40 amino acids typically bracketed by glycine–histidine (GH) and tryptophan–aspartic acid (WD), which can promote the formation of heterotrimeric or polyprotein complexes. Members of the WD-repeat family are involved in a variety of cellular processes, including signal transduction, transcription regulation, cell cycle progression, autophagy, and apoptosis.14,15 WDR12, as a component of the PeboW complex, participates in ribosome biogenesis and cell proliferation.3 Previous studies have suggested that WDR12 was related to cardiovascular diseases, including myocardial infarction and coronary artery disease.16 However, the role of WDR12 in human tumors has not been explored. Herein, we have demonstrated that WDR12 was aberrantly overexpressed in human HCC patients and cell lines. Significantly, WDR12 expression was negatively correlated with overall and disease-free survival in patients with HCC. Knockdown of WDR12 by shRNAs revealed that WDR12 supports HCC development by promoting the proliferation and migration of HCC cells. Herein, we first demonstrated the function of WDR12 in supporting HCC development. This finding may provide novel insights into the molecular function of WDR12 in cancer. Importantly, previous studies suggested that the WD-repeat domain has a structurally diverse pocket that may be pharmaceutically acceptable.15 Thus, our findings may be helpful in designing new therapeutic strategies for patients with HCC.
Most WD repeat proteins seem to be regulatory and perform crucial roles in signal transduction. Many studies have shown that there are many WD-repeat proteins involved in signal transduction in many human diseases, including cancer.17 Recently, Chen et al18 showed that WDR79 regulates the ubiquitination and degradation of UHRF1 and thus mediates the proliferation of nonsmall-cell lung cancer cells. In addition, WDR5 was shown to promote the proliferation of gastric cancer cells by mediating the expression of cyclin D1.19 Another study showed that WDR5 knockdown significantly inhibits the proliferation, invasion, and migration of head neck squamous cell carcinoma cells.20 Collectively, these studies suggest that WD-repeat proteins play significant roles in the proliferation and motility of cancer cells by regulating a variety of signaling pathways. Nevertheless, the molecular mechanism of the function of WDR12 in promoting different aspects of tumor malignancy remains poorly determined. A new discovery in our research is that WDR12 promotes the activation of Akt-mTOR-S6K1 signaling. A previous study showed that WD-repeat propeller-FYVE protein (ProF) is a binding partner of the serine/threonine protein kinase Akt. ProF consists of seven WD repeats, acting as a protein–protein binding platform, whereas the FYVE domain interacts with Akt.21,22 Given that WDR12 does not possess any enzymatic activity, we speculate that WDR12 may act as a protein–protein binding platform and promote interactions between Akt and other FYVE-containing kinases. Thus, the WD-repeat propeller domains of WDR12 may play crucial roles in promoting Akt and mTOR phosphorylation. Further studies are required to confirm the mechanism of how WDR12 regulates Akt and mTOR activation both in vitro and in vivo.
Conclusion
Altogether, we have revealed a new effect of WDR12 on maintaining the proliferation and migration of HCC cells by promoting Akt-mTOR-S6K1 signaling. WDR12 is highly enriched in functional HCC cells and might be an ideal therapeutic target for treating human HCC. The function of WDR12 in supporting cancer development in vivo in HCC or other solid tumors warrants further study.
Acknowledgments
This work was supported by the National Natural Science Foundation of China [grant numbers 31501146 and 81600128] and the Natural Science Foundation of Shandong Province [grant numbers BS2015SW024 and BS2015YY013].
Author contributions
YY contributed to performing experiments, interpreting data, and writing the manuscript. LZ, RZ, and QZ contributed to in silico analysis. ML contributed to experimental design and writing the manuscript. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
Dutta R, Mahato RI. Recent advances in hepatocellular carcinoma therapy. Pharmacol Ther. 2017;173:106–117. | ||
Hölzel M, Rohrmoser M, Schlee M, et al. Mammalian WDR12 is a novel member of the Pes1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J Cell Biol. 2005;170(3):367–378. | ||
Romes EM, Sobhany M, Stanley RE. The crystal structure of the ubiquitin-like domain of ribosome assembly factor Ytm1 and characterization of its interaction with the AAA-ATPase midasin. J Biol Chem. 2016;291(2):882–893. | ||
Rohrmoser M, Hölzel M, Grimm T, et al. Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol Cell Biol. 2007;27(10):3682–3694. | ||
Lewinska A, Bednarz D, Adamczyk-Grochala J, Wnuk M. Phytochemical-induced nucleolar stress results in the inhibition of breast cancer cell proliferation. Redox Biol. 2017;12:469–482. | ||
Rose BA, Force T, Wang Y. Mitogen-activated protein kinase signaling in the heart: angels versus demons in a heart-breaking tale. Physiol Rev. 2010;90(4):1507–1546. | ||
Willis MS, Patterson C. Hold me tight: Role of the heat shock protein family of chaperones in cardiac disease. Circulation. 2010;122(17):1740–1751. | ||
Lerch-Gaggl A, Haque J, Li J, et al. Pescadillo is essential for nucleolar assembly, ribosome biogenesis, and mammalian cell proliferation. J Biol Chem. 2002;277(47):45347–45355. | ||
Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. | ||
Chen C, Yin Y, Li C, et al. ACER3 supports development of acute myeloid leukemia. Biochem Biophys Res Commun. 2016;478(1):33–38. | ||
Yin Y, Hua H, Li M, et al. mTORC2 promotes type I insulin-like growth factor receptor and insulin receptor activation through the tyrosine kinase activity of mTOR. Cell Res. 2016;26(1):46–65. | ||
Li M, Jin C, Xu M, et al. Bifunctional enzyme ATIC promotes propagation of hepatocellular carcinoma by regulating AMPK-mTOR-S6K1 signaling. Cell Commun Signal. 2017;15(1):52. | ||
Stirnimann CU, Petsalaki E, Russell RB, Müller CW. WD40 proteins propel cellular networks. Trends Biochem Sci. 2010;35(10):565–574. | ||
Schapira M, Tyers M, Torrent M, Arrowsmith CH. WD40 repeat domain proteins: a novel target class? Nat Rev Drug Discov. 2017;16(11):773–786. | ||
Myocardial Infarction Genetics Consortium, Kathiresan S, Voight BF, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41(3):762–341. | ||
Li D, Roberts R. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell Mol Life Sci. 2001;58(14):2085–2097. | ||
Chen J, Sheng X, Ma H, et al. WDR79 mediates the proliferation of non-small cell lung cancer cells by regulating the stability of UHRF1. J Cell Mol Med. 2018;22(5):2856–2864. | ||
Sun W, Guo F, Liu M. Up-regulated WDR5 promotes gastric cancer formation by induced cyclin D1 expression. J Cell Biochem. 2018;119(4):3304–3316. | ||
Wu Y, Diao P, Li Z, et al. Overexpression of WD repeat domain 5 associates with aggressive clinicopathological features and unfavorable prognosis in head neck squamous cell carcinoma. J Oral Pathol Med. 2018;47(5):502–510. | ||
Fritzius T, Moelling K, Akt-. Akt- and Foxo1-interacting WD-repeat-FYVE protein promotes adipogenesis. Embo J. 2008;27(9):1399–1410. | ||
Fritzius T, Burkard G, Haas E, et al. A WD-FYVE protein binds to the kinases Akt and PKCzeta/lambda. Biochem J. 2006;399(1):9–20. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.