Back to Journals » Cancer Management and Research » Volume 13
Identification of TRIM56 as a Potential Biomarker for Lung Adenocarcinoma
Received 28 October 2020
Accepted for publication 17 February 2021
Published 4 March 2021 Volume 2021:13 Pages 2201—2213
DOI https://doi.org/10.2147/CMAR.S288111
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Harikrishna Nakshatri
Kun Lu, Yingli Sui, Lin Fu
Institute of Chronic Disease, School of Basic Medicine, Qingdao University, Qingdao, Shandong, People’s Republic of China
Correspondence: Lin Fu
Institute of Chronic Disease, School of Basic Medicine, Qingdao University, No. 308 Ningxia Road, Qingdao City, Shandong Province, 266071, People’s Republic of China
Tel +86 138 1804 8288
Fax +86 532-85953085
Email [email protected]
Background: Lung adenocarcinoma (LUAD) is the primary subtype of human lung cancer. The effectiveness of treatment and long-term survival of patients with LUAD are current suboptimal. Tripartite motif containing 56 (TRIM56) is a member of the TRIM protein family that have functions predominantly in immunity and cancer.
Purpose: To investigate the expression of TRIM56 in LUAD, and explore the potential regulatory role of TRIM56 in the invasion and migration of LUAD cells.
Methods: The Gene Expression Omnibus datasets and The Cancer Genome Atlas-LUAD cohort were used to analyze the mRNA expression of TRIM56 in LUAD. The differential expression profiles of miRNAs associated with TRIM56 were obtained from The Cancer Genome Atlas-LUAD cohort. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses were performed to determine the principal functions of miRNAs and interacting proteins. Transwell and wound healing were used to detect the effect of overexpression of TRIM56 on the invasion and migration of LUAD cells.
Results: The expression of TRIM56 was decreased in LUAD and associated with poor prognosis. We determined the genome copy number, negatively correlated miRNA and potential transcription factors of TRIM56, and conducted enrichment analysis. Among them, hsa-mir-542 and hsa-mir-627 were the most likely to inhibit the expression of TRIM56. We also predicted the interacting proteins and potential ubiquitination substrate of TRIM56. Finally, we demonstrated that overexpression of TRIM56 inhibits the invasion and migration of LUAD cells.
Conclusion: This study is the first to analyze the expression of TRIM56 and its inhibitory effect on the invasion and migration of LUAD. This evidence provides a new direction for further study of the reasons for the low expression of TRIM56 in LUAD and its regulatory mechanism.
Keywords: TRIM56, lung adenocarcinoma, bioinformatics analysis, invasion, migration
Introduction
Lung cancer is one of the most common tumors and associated with the highest rates of morbidity and mortality worldwide.1 In 2017, the death toll due to lung cancer exceeded the total caused by prostate, colorectal, brain, and breast cancer. Almost one-quarter of cancer-related deaths are due to lung cancer.2 Epidemiological statistics have shown that lung adenocarcinoma (LUAD) has become the primary subtype of lung cancer.2 Based on its characteristics, LUAD is prone to recurrence and metastasis; in addition, the therapeutic effect in patients with LUAD is currently suboptimal, and their long-term survival rate remains low.3 In general, the occurrence and development of tumors is a complex, multi-stage process, involving numerous signal transduction pathways, and a variety of factors in vitro and in vivo. Therefore, investigation of the abnormally altered genes in LUAD is crucial to further elucidate the relevant mechanisms involved in its occurrence and development.
The human genome encodes more than 70 members of the tripartite motif containing (TRIM) family. In previous reports, almost all TRIM proteins exhibited E3 ubiquitin ligase activity depending on their RING finger domain, which mediates the ubiquitination of various substrates and themselves.4 TRIM proteins are involved in various cell processes, including differentiation, proliferation, apoptosis, and morphogenesis. Moreover, evidence on its role in immune signal transduction and antiviral functions is emerging.5,6
TRIM56 is an important member of the TRIM family. At present, research on TRIM56 mainly focuses on the antiviral activity TRIM56 can exert by mediating the K63 ubiquitin link of stimulator of interferon genes (STING) (a type I interferon stimulating gene), as well as the induction process of type I interferon by double-stranded DNA of viruses and other pathogens.7 TRIM56 can also activate the toll like receptor 3 (TLR3) signaling pathway and promote the production of interferon. Of note, this process is independent of its E3 ubiquitin ligase activity.8,9 In addition, TRIM56 can also inhibit the RNA replication of influenza viruses A and B through its specific C-terminal functional domain.10
However, there are few studies on TRIM56 in tumors. TRIM56 inhibits the proliferation of multiple myeloma cells and induces apoptosis.11 In ovarian cancer, TRIM56 can mediate the K48 ubiquitin connection of vimentin to promote its degradation, thus inhibiting the development of ovarian cancer.12 Interestingly, in breast cancer, TRIM56 mediates K63 ubiquitination with estrogen receptor-alpha (ER-α), upregulates the protein stability of ER-α, and promotes the proliferation of breast cancer cells.13 Therefore, the role of TRIM56 in tumors remains unclear, and alterations in its expression and function in lung cancer have not been reported.In our study, we will explore the relationship between abnormal expression of TRIM56 and lung adenocarcinoma through bioinformatics analysis, and verify that TRIM56 overexpression suppress the invasion and migration of lung adenocarcinoma cells.
Materials and Methods
Cells Culture and Plasmids
H1299 cells were obtained from the American Type Culture Collection (Manassas, VA, USA), while 293T cells for cell transfection purchased from Gene Pharma (China). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; high glucose) supplemented with 10% fetal bovine serum (Bioind, USA) and 1% penicillin/streptomycin (Solarbio, Beijing, China) in a humidified atmosphere with 5% CO2 at 37°C. The transfection reagent Lipofectamine 2000 was purchased from Vigorous Biotechnology (China). Plasmids for cell transfection were provided and constructed by a laboratory.
Western Blotting
H1299 cells were collected and lysed with radioimmunoprecipitation assay buffer (Beyotime, Shanghai, China). Total cellular protein was collected using 5 × sodium dodecyl sulfate sample buffer and denatured for 10 min at 100°C. Proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrotransferred to polyvinylidene difluoride membranes. After blocking with 5% skimmed milk (weight/volume), the polyvinylidene difluoride membranes were incubated with the following antibodies: mouse monoclonal anti-Myc antibody (Cell Signaling Technology, Danvers, MA, USA) and monoclonal rabbit anti-glyceraldehyde-3-phosphate dehydrogenase (Proteintech, Rosemont, IL, USA). Subsequently, the membranes were incubated with mouse and rabbit secondary antibodies for 1.5 h. A highly sensitive substrate (Millipore, Billerica, MA, USA) was used to detect specific bands on autoradiographic film.
Lentivirus Packaging and Infection
pCDH-Myc-TRIM56 or pCDH empty vector and two packaging plasmids (delta 8.9, pLP-VSVG) were stably transfected into 293T cells using the Lipofectamine 2000 transfection reagent (Thermo Scientific, Waltham, MA, USA). The concentration of lentivirus was determined after 48 h and 72 h using filtered virus supernatant. H1299 cells were infected with virus concentrate (5 μL) for 48 h, and the stability of transfection was assessed using puromycin. Polybrene (8 μg/mL) was used to infect cells with viruses.
UALCAN Analysis
UALCAN (http://ualcan.path.uab.edu) is an open interactive website. The website includes RNA sequencing, clinical research, and DNA methylation data regarding 31 types of cancer, which are available in The Cancer Genome Atlas (TCGA) database.14 UALCAN can analyze the relative expression of genes in tumors and normal samples, as well as various tumor subgroups based on clinicopathological characteristics. We used UALCAN to analyze the relationship between the expression of TRIM56 and the clinical characteristics of LUAD, including the patient’s age, race, sex, lymph node metastasis, tumor stage, and tumor protein p53 (TP53) mutation. Furthermore, we analyzed the relationship between promoter methylation and TRIM56 expression in patients with LUAD.
Gene Expression Profiling Interactive Analysis (GEPIA) Analysis
GEPIA (http://gepia.cancer-pku.cn/) is a web-based tool,15 providing customizable analysis functions. These functions include gene expression analysis in tumors and normal tissues, multi-gene expression correlation analysis, and patient survival analysis. The main data sources of GEPIA are TCGA and Genotype-Tissue Expression databases. We used GEPIA to analyze the expression of TRIM56 in three subtypes of LUAD, namely terminal respiratory unit, proximal hyperplasia, and proximal inflammatory.
LinkedOmics Analysis
LinkedOmics is a publicly available portal (http://www.LinkedOmics.org/login.php) that includes multi-omics data regarding 32 types of cancer, which are available in TCGA.16 We used the LinkFinder module of LinkedOmics to explore the differentially expressed miRNA associated with TRIM56 in the TCGA-LUAD cohort. Pearson correlation coefficients were determined to explore the negative correlations with TRIM56. All results are graphically displayed in the volcano and heat map. A false discover ratio <0.05 was set as the threshold.
cBioPortal Analysis
The cBioPortal database (http://cbioportal.org) is an open-source tool for the interactive exploration of multi-dimensional cancer genomics datasets and currently contains evidence obtained from 308 cancer studies.17,18 We used the cBioPortal to analyze TRIM56 copy number alteration (CNA), mutations, and changes in DNA methylation using data from TCGA-LUAD.
Functional Enrichment (FunRich) Analysis Tool
We performed enrichment analysis of negatively correlated miRNAs using the FunRich tool (http://www.funrich.org/). FunRich is a stand-alone software tool used mainly for functional enrichment and interaction network analysis of genes and proteins.19,20 We used FunRich to analyze the biological process, cellular component, molecular function, and biological pathways.
The Kaplan–Meier Plotter
The prognostic value of the expression of TRIM56 in GSE31210 and negatively correlated miRNAs was evaluated using the online database Kaplan–Meier Plotter (http://www.kmplot.com).21,22 To analyze the survival of patients with LUAD, patient samples were classified into two groups according to their median expression (high versus low expression). A Kaplan–Meier survival plot was produced, and the results included hazard ratios with 95% confidence intervals and log-rank p-values.
Search Tool for the Retrieval of Interacting Genes (STRING)
STRING is a protein–protein interaction database, involving known and predicted data (https://string-db.org/). The protein interaction data available in STRING include direct (physical) and indirect (functional) associations; they stem from computational prediction, knowledge transfer between organisms, and interactions aggregated from other (primary) databases.23,24 We used STRING to analyze proteins interacting with TRIM56 and the enrichment analysis data.
Transwell Assays
Corning Matrigel was purchased from Becton, Dickinson and Company (USA). H1299 cells were stably transfected with pCDH-Myc-TRIM56 or pCDH empty vector, and seeded at a density of 1×104 cells per chamber in 24-well plates. DMEM medium with 0.1% bovine serum albumin was placed in the upper chamber. DMEM medium with 25% fetal bovine serum was added in the lower chamber.
Wound Healing Assays
H1299 cells were stably transfected with pCDH-Myc-TRIM56 or pCDH empty vector, and seeded at a density of 1×105 cells per well in six-well plates. After drawing lines for 0 h, the DMEM high glucose medium containing 100× penicillin-streptomycin solution was replaced.
qRT-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA). First strand cDNA was synthesized subsequently using the Fast Quant RT Kit (with gDNase) (TOYOBO,Japan) according to the product manual. Quantitative polymerase chain reaction (qPCR) was performed in three replicate wells on an ABI 7500 Real-Time PCR System (ThermoFisher Scientific, Waltham, MA) using SuperReal PreMix Plus (SYBR Green) (Tiangen Biotech, Beijing, China). GAPDH, forward:CATGAGAAGTATGACAACAGCCT; reverse: AGTCCTTCCACGATACAAAG. TRIM56,forward:GATCAGCTCTGGCTAGTTCTCACA;reverse:CTAGTCTTCTGAGGCCTTGGGC.The relative expression levels of TRIM56 were calculated using the 2−ΔΔCT method and GAPDH was used as an internal control. Each experiment was repeated three times.
Statistical Analysis
The cut-off values for TRIM56 expression were determined by their median values. All analyses, including the t-test and correlation analysis were performed by GraphPad Prism. In our analysis, P values less than 0.05 were considered significant.
Results
TRIM56 Was Downregulated in LUAD and Associated with Poor Prognosis
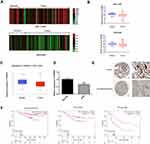
To determine the expression levels of TRIM56 in LUAD, we first analyzed its mRNA expression by searching the open databases Gene Expression Omnibus (GEO) and TCGA. The results showed that the expression of TRIM56 in LUAD samples was lower than that observed in normal tissues in GEO datasets (GSE117049 and GSE43458) (Figure 1A and B). Consistent with the GEO results, TRIM56 showed low expression in LUAD tissue from the TCGA-LUAD cohort (Figure 1C). To verify the mRNA expression of TRIM56 in lung adenocarcinoma cells and normal lung cells, we performed qRT-PCR to detect the TRIM56 mRNA levels. We found that the mRNA levels of TRIM56 was down-regulated in A549 cells compared to Beas-2B cells (Figure 1D). In addition, representative images from The Human Protein Atlas (https://www.proteinatlas.org/)25 demonstrated that TRIM56 protein expression was lower in LUAD samples compared with normal lung tissues (Figure 1E).
Next, we evaluated the prognostic value of TRIM56 in LUAD. We used the GSE31210 dataset to analyze the relationship between the mRNA expression of TRIM56 and overall, pre-tumor, and late-stage tumor survival. The analysis results showed that the downregulation of TRIM56 was associated with poor prognosis of LUAD in terms of overall, pre-tumor, and late-stage tumor survivals (Figure 1F). Therefore, TRIM56 is poorly expressed in LUAD and related to prognosis.
In addition, we analyzed the relationship between the expression of TRIM56 and the clinicopathological features of LUAD. Notably, the expression levels of TRIM56 did not show a difference between groups classified by sex, smoking habit, nodal metastasis status, age, and tumor stage (Supplementary Figure 1A–E). The expression of TRIM56 was significantly higher in Caucasian than African-American patients (Supplementary Figure 1F). In addition, its expression was higher in the TP53 mutation group versus the TP53 non-mutation group (Supplementary Figure 1G). TRIM56 was downregulated in the three subtypes of LUAD, namely proximal inflammatory, proximal proliferative, and terminal respiratory unit (Supplementary Figure 1H–J).26,27 The reduced expression of TRIM56 was more pronounced in the proximal proliferative subtype (Supplementary Figure 1I). This subtype is associated with LUAD metastases to the brain,28 suggesting that TRIM56 may affect tumor migration and invasion.
The Expression of TRIM56 Was Regulated by DNA CNA in LUAD
In this study, we used RNA sequencing, copy number, mutation, and DNA methylation data available in TCGA-LUAD to investigate the potential mechanism of TRIM56 dysregulation. Firstly, we detected the genetic changes of TRIM56 in the TCGA-LUAD cohort using the cancer genomics data available in cBioportal. Among 816 LUAD samples for which sequencing and DNA CNA data were available, changes in the DNA sequence of TRIM56 were observed in 27 (4%) samples (Figure 2A); the main type of gene mutation was gene amplification. Subsequently, we used the UCSC genome browser database (https://genome.ucsc.edu) to simultaneously detect mRNA expression, CNA, and DNA methylation of TRIM56 in a total of 611 patients (Figure 2B). Next, we used the TCGA-LUAD dataset in cBioportal to analyze the effect of CNA on the expression of TRIM56 (Figure 2C). The results showed a significant correlation between DNA deletion and the downregulation of TRIM56 expression. Furthermore, the probability of a missense mutation (Missense) and a truncating mutation (Truncating) in TRIM56 was 1.86% and 0.16%, respectively. However, the mutation of TRIM56 did not affect its mRNA levels (Figure 2D). Although regression analysis showed that the expression of TRIM56 was negatively correlated with its methylation status (Figure 2E), the level of TRIM56 promoter methylation in LUAD samples was lower than that recorded in normal tissues (Figure 2F). Therefore, we hypothesized that DNA methylation does not play a major role in the regulatory mechanism for the expression of TRIM56.
miRNAs and Transcription Factors May Regulate the Expression of TRIM56 in LUAD
In addition to changes in its DNA sequence, we hypothesized that TRIM56 may be regulated by miRNAs and transcription factors. Firstly, we used LinkedOmics to identify the miRNAs that are negatively related to TRIM56 expression. The volcano map is shown in Figure 3A. The red and green dots represent miRNAs that are positively (n=407) and negatively (n=403) correlated with TRIM56 expression, respectively. Typically, miRNAs suppress the expression of their target genes. In this analysis, we explored the top 50 miRNAs most negatively correlated with TRIM56 expression (Figure 3B).
Next, we used the FunRich tool to perform enrichment analysis on the target genes of the top 30 miRNAs that were screened. The results showed that the pathways for the enrichment of target genes of these miRNAs include the glypican pathway, proteoglycan syndecan-mediated signaling events, interferon-gamma (IFNG) pathway, alpha9 beta1 integrin signaling events, ErbB receptor signaling network, and vascular endothelial growth factor (VEGF) and VEFG receptor (VEGFR) signaling network (Figure 3C). The main enrichment biological processes of these miRNA target genes are signal transduction, cell communication, transport, regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolism, regulation of cell growth, regulation of gene expression, and epigenetic regulation (Figure 3D). The main enriched molecular functions of these miRNA target genes are transcription factor activity, ubiquitin-specific protease activity, protein serine/threonine kinase activity, GTPase activity, receptor binding, and receptor signaling complex scaffold activity (Figure 3E). The target genes of these miRNAs are mainly located in the nucleus, cytoplasm, lysosome, endosome, Golgi apparatus, and receptor complex (Figure 3F). In addition, we analyzed the expression and prognostic value of the top 20 miRNAs, and found that hsa-mir-542 and hsa-mir-627 were abnormally overexpressed and related to poor prognosis of LUAD (Figure 3G and H; Table 1). These results imply that these miRNAs may affect the prognosis of LUAD by regulating the expression of TRIM56.
 |
Table 1 Overall Survival of miRNA That Target TRIM56 in LUAD |
The regulation of TRIM56 expression by transcription factors has not been previously reported. Thus, we analyzed the potential transcription factors of TRIM56. Firstly, we retrieved the TRIM56 promoter sequence through the UCSC Genome Browser, and subsequently used PROMO and ConTra V3 to identify possible transcription factors (Table 2). There were 22 and four potential transcription factors identified from PROMO and ConTra v3, respectively. We also examined the correlation between the expression of these transcription factors and that of TRIM56 in LUAD. TRIM56 was significantly correlated with the following transcription factors: transcription factor II–I (TFII-I), RXR-alpha, AP-2 alpha, Pax-5, c-Ets-1, zinc finger and BTB domain containing 7B (ZBTB7B), and SP2 (Table 2). Therefore, these transcription factors may regulate the expression of TRIM56 in LUAD.
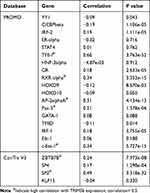 |
Table 2 Transcription Factor of TRIM56 in PROMO and ConTra V3 |
Gene Ontology (GO) Functional Annotation and Pathway Enrichment of Proteins Interacting with TRIM56
As an E3 ubiquitin ligase, TRIM56 functions by binding to other proteins. Proteome analysis revealed 20 proteins interacting with TRIM56; these proteins were displayed in a physical interaction network using STRING and Cytoscape (Figure 4A). The most reliable proteins were LSM2, TRIM38, Mab-21 domain containing 1 (MB21D1), T cell receptor associated transmembrane adaptor 1 (TRAT1), transmembrane protein 137 (TMEM137), and TRIM32 (Figure 4A).
Next, we performed GO and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis of the 20 aforementioned proteins interacting with TRIM56. These proteins are involved in numerous biological processes, such as negative regulation of gene expression, nuclear-transcribed mRNA catabolic process, nonsense-mediated decay, protein targeting, signal recognition particle-dependent cotranslational protein targeting to membrane, mRNA metabolic process, GO: 0044265cellular macromolecule catabolic process, and nuclear-transcribed mRNA catabolic process (Figure 4B). Moreover, we noticed significant enrichment of functions corrected with U6 snRNA binding, structural constituent of ribosome, RNA binding, translation initiation factor binding, and rRNA binding (Figure 4C). These proteins are mainly localized to the U6 snRNP, ribonucleoprotein complex, spliceosomal tri-snRNP complex, U4/U6 x U5 tri-snRNP complex, Lsm1-7-Pat1 complex, and ribosome (Figure 4D). The Kyoto Encyclopedia of Genes and Genomes pathways in which these proteins are mainly involved are (hsa03010) ribosome, (hsa03018) RNA degradation, (hsa03040) spliceosome, and (hsa04623) cytosolic DNA-sensing pathway (Figure 4E).
The predicted ubiquitination substrates of TRIM56 are shown in Table 3. The expression of TP53, H2A histone family member X (H2AFX), lymphoid enhancer binding factor 1 (LEF1), and hepatocyte growth factor-regulated tyrosine kinase substrate (HGS) is abnormal in LUAD. In summary, we analyzed the proteins interacting with TRIM56 and predicted the potential ubiquitination substrates of TRIM56.
 |
Table 3 Ubiquitination Substrates of TRIM56 in LUAD from UbiBrowser |
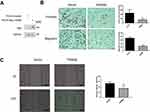
Overexpression of TRIM56 Suppressed the Invasion and Migration of LUAD Cells
To determine the effect of TRIM56 on the invasive and migratory ability of LUAD cells, we stably expressed Myc-TRIM56 in H1299 cells via a lentiviral system. As shown in Figure 5A, the expression of TRIM56 was detected by Western blotting. Next, we examined the invasive and migratory ability of pCDH-TRIM56 H1299 cells using Transwell assays (Figure 5B). We found that the invasion and migration of H1299 cells overexpressing TRIM56 were significantly reduced than those observed in the control group. Consistent with these results, wound healing experiments showed that the migratory ability of pCDH-TRIM56 H1299 cells was significantly downregulated (Figure 5C). Therefore, overexpression of TRIM56 may suppress the invasion and migration of LUAD cells.
Discussion
TRIM56 catalyzes the transfer of ubiquitin molecules to specific substrates.4 TRIM56 was localized in the cytoplasm and failed to interact with double-stranded DNA. However, it is responsible for the induction of type I IFN by promoting the function of STING through ubiquitination.7 In addition, TRIM56 can activate TLR3 antiviral signaling pathway-induced interferon.9 These findings confirm that TRIM56 plays an important role in antiviral and immune regulation. Previous studies have shown that the expression of TRIM56 is reduced in a variety of human cancers, including multiple myeloma11 and ovarian cancer.29 In these tumors, the low expression of TRIM56 is associated with tumor development, metastasis, and poor prognosis. However, TRIM56 can promote breast cancer through ubiquitination and stabilize ER-α.13 In summary, TRIM56 plays various roles in different tumors. This is the first study to demonstrate that the expression of TRIM56 in LUAD was decreased and related to poor prognosis. The migration and invasion experiments in vitro were verified that TRIM56 overexpression suppress the invasion and migration of lung adenocarcinoma cells. It provides a basis for further in vivo experiments.
In general, genetic and epigenetic alterations can regulate the tumor phenotype and patient survival.30 In this study, the potential mechanism underlying the dysregulation of TRIM56 was further investigated using deep sequencing data from the TCGA-LUAD cohort. The present findings showed that upregulated TRIM56 expression was significantly associated with DNA copy deletion rather than DNA mutations and methylation. Moreover, TRIM56 expression was negatively correlated with DNA methylation. Nevertheless, the methylation of the promoter of TRIM56 in patients with LUAD was reduced, implying the presence of an unknown mechanism for the regulation of TRIM56 methylation in LUAD.
Previous research found that hsa-mir-9 promoted the development and progression of multiple myeloma by regulating the TRIM56/NF-κB pathway.31 In this study, we found two miRNAs negatively correlated with TRIM56 expression and associated with poor prognosis of LUAD, namely hsa-mir-542 and hsa-mir-627. These miRNAs exert important regulatory effects on tumors. Hsa-mir-542 can be used as a non-invasive blood biomarker for tumor monitoring and prognosis prediction in patients with osteosarcoma.32 Moreover, it can target CDK14 to inhibit the development of ovarian cancer,33 and inhibit the growth and invasion of colon cancer cells through PI3K/AKT/survivin signaling.34 Hence, these two miRNAs may affect the prognosis of LUAD by inhibiting the expression of TRIM56.
The enrichment analysis of proteins interacting with TRIM56 showed that TRIM56 may be involved in numerous biological processes of mRNA splicing and ribosomes. During mRNA splicing, pre-mRNA is assembled into a large, dynamic spliceosome. This process requires the participation of five small nuclear RNAs (U1, U2, U4, U5, and U6) and more than 50 protein components.35 The splicing and maturation of mRNA are essential for cell growth and development. Therefore, it is possible that TRIM56 regulates tumors by regulating gene splicing and maturation.
Conclusion
In this study, we analyzed the expression of TRIM56 in LUAD using bioinformatics. The results demonstrated that the low expression of TRIM56 was related to poor prognosis of LUAD. In addition, we discussed the change in gene copy number and the regulatory effects of miRNA and transcription factors on the mRNA expression levels of TRIM56. Furthermore, we predicted the proteins interacting with TRIM56 and analyzed their enrichment. Finally, our experimental results showed that TRIM56 can inhibit the invasion and migration of LUAD. In summary, the present research showed, for the first time, that TRIM56 is a potential tumor suppressor. This evidence provides a new direction for analyzing the reasons for the low expression of TRIM56 in LUAD and its regulatory mechanism involved in tumor migration and invasion.
Acknowledgments
We appreciated Dr. Likun Zhuang and Dr. Shousheng Liu for providing the experimental apparatus for the experiments involved in the manuscript.
Funding
This project is supported by grants from the National Natural Science Foundation of China [81702743] and the China Postdoctoral Science Foundation [2018M640612, 2019T120568]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
3. Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–289. doi:10.3322/caac.21349
4. Zhan W, Zhang S. TRIM proteins in lung cancer: mechanisms, biomarkers and therapeutic targets. Life Sci. 2021;268:118985. doi:10.1016/j.lfs.2020.118985
5. Wang HT, Hur S. Substrate recognition by TRIM and TRIM-like proteins in innate immunity. Semin Cell Dev Biol. 2020. doi:10.1016/j.semcdb.2020.09.013
6. Ozato K, Shin DM, Chang TH, Morse HC. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8(11):849–860. doi:10.1038/nri2413
7. Tsuchida T, Zou J, Saitoh T, et al. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33(5):765–776. doi:10.1016/j.immuni.2010.10.013
8. Wang J, Liu B, Wang N, Lee YM, Liu C, Li K. TRIM56 is a virus- and interferon-inducible E3 ubiquitin ligase that restricts pestivirus infection. J Virol. 2011;85(8):3733–3745. doi:10.1128/JVI.02546-10
9. Shen Y, Li NL, Wang J, Liu B, Lester S, Li K. TRIM56 is an essential component of the TLR3 antiviral signaling pathway. J Biol Chem. 2012;287(43):36404–36413. doi:10.1074/jbc.M112.397075
10. Liu B, Li NL, Shen Y, et al. The C-terminal tail of TRIM56 dictates antiviral restriction of influenza A and B viruses by impeding viral RNA synthesis. J Virol. 2016;90(9):4369–4382. doi:10.1128/JVI.03172-15
11. Chen Y, Zhao J, Li D, et al. TRIM56 suppresses multiple myeloma progression by activating TLR3/TRIF signaling. Yonsei Med J. 2018;59(1):43–50. doi:10.3349/ymj.2018.59.1.43
12. Zhao L, Zhang P, Su XJ, Zhang B. The ubiquitin ligase TRIM56 inhibits ovarian cancer progression by targeting vimentin. J Cell Physiol. 2018;233(3):2420–2425. doi:10.1002/jcp.26114
13. Xue M, Zhang K, Mu K, et al. Regulation of estrogen signaling and breast cancer proliferation by an ubiquitin ligase TRIM56. Oncogenesis. 2019;8(5):30. doi:10.1038/s41389-019-0139-x
14. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi:10.1016/j.neo.2017.05.002
15. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–w102. doi:10.1093/nar/gkx247
16. Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–d63. doi:10.1093/nar/gkx1090
17. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi:10.1126/scisignal.2004088
18. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi:10.1158/2159-8290.CD-12-0095
19. Pathan M, Keerthikumar S, Ang CS, et al. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15(15):2597–2601. doi:10.1002/pmic.201400515
20. Pathan M, Keerthikumar S, Chisanga D, et al. A novel community driven software for functional enrichment analysis of extracellular vesicles data. J Extracell Vesicles. 2017;6(1):1321455. doi:10.1080/20013078.2017.1321455
21. Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. doi:10.1371/journal.pone.0082241
22. Á N, Lánczky A, Menyhárt O, Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8(1):9227. doi:10.1038/s41598-018-27521-y
23. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–d13. doi:10.1093/nar/gky1131
24. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–d68. doi:10.1093/nar/gkw937
25. Uhlén M, Fagerberg L, Hallström BM, et al. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi:10.1126/science.1260419
26. Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi:10.1038/nature13385
27. Wilkerson MD, Yin X, Walter V, et al. Differential pathogenesis of lung adenocarcinoma subtypes involving sequence mutations, copy number, chromosomal instability, and methylation. PLoS One. 2012;7(5):e36530. doi:10.1371/journal.pone.0036530
28. Faruki H, Mayhew GM, Serody JS, Hayes DN, Perou CM, Lai-Goldman M. Lung adenocarcinoma and squamous cell carcinoma gene expression subtypes demonstrate significant differences in tumor immune landscape. J Thorac Oncol. 2017;12(6):943–953. doi:10.1016/j.jtho.2017.03.010
29. Zhao L, Wang ZG, Zhang P, Yu XF, Su XJ. Poly r(C) binding protein 1 regulates posttranscriptional expression of the ubiquitin ligase TRIM56 in ovarian cancer. IUBMB Life. 2019;71(2):177–182. doi:10.1002/iub.1948
30. Huang N, Lin W, Shi X, Tao T. STK24 expression is modulated by DNA copy number/methylation in lung adenocarcinoma and predicts poor survival. Future Oncol. 2018;14(22):2253–2263. doi:10.2217/fon-2018-0126
31. Huang G, Liu X, Zhao X, et al. MiR-9 promotes multiple myeloma progression by regulating TRIM56/NF-kappaB pathway. Cell Biol Int. 2019;43(11):1223–1233. doi:10.1002/cbin.11104
32. Li Q, Song S, Ni G, Li Y, Wang X. Serum miR-542-3p as a prognostic biomarker in osteosarcoma. Cancer Biomarkers. 2018;21(3):521–526. doi:10.3233/CBM-170255
33. Li J, Shao W, Feng H. MiR-542-3p, a microRNA targeting CDK14, suppresses cell proliferation, invasiveness, and tumorigenesis of epithelial ovarian cancer. Biomed Pharmacother. 2019;110:850–856. doi:10.1016/j.biopha.2018.11.104
34. Yang C, Wang MH, Zhou JD, Chi Q. Upregulation of miR-542-3p inhibits the growth and invasion of human colon cancer cells through PI3K/AKT/survivin signaling. Oncol Rep. 2017;38(6):3545–3553. doi:10.3892/or.2017.6054
35. Yan C, Wan R, Shi Y. Molecular mechanisms of pre-mRNA splicing through structural biology of the spliceosome. Cold Spring Harb Perspect Biol. 2019;11(1):a032409. doi:10.1101/cshperspect.a032409
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.