Back to Journals » International Journal of General Medicine » Volume 14
Identification of Three Potential circRNA Biomarkers of Polycystic Ovary Syndrome by Bioinformatics Analysis and Validation
Authors Huang P, Du S, Lin Y, Huang Z, Li H, Chen G, Chen S, Chen Q, Da L , Shi H, Wei W, Yang L, Sun Y, Zheng B
Received 16 June 2021
Accepted for publication 26 August 2021
Published 22 September 2021 Volume 2021:14 Pages 5959—5968
DOI https://doi.org/10.2147/IJGM.S324126
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Pengyu Huang,* Shengrong Du,* Yunhong Lin,* Zhiqing Huang, Haiyan Li, Gangxin Chen, Suzhu Chen, Qingfen Chen, Lincui Da, Hang Shi, Wei Wei, Lei Yang, Yan Sun, Beihong Zheng
Reproductive Medicine Center, Fujian Maternity and Child Health Hospital, Affiliated Hospital of Fujian Medical University, Fuzhou, 350001, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yan Sun; Beihong Zheng Email [email protected]; [email protected]
Objective: It is well known that circRNAs are closely involved in the progression of various diseases. However, their functions and potential regulatory mechanisms in polycystic ovary syndrome (PCOS) remain largely unknown. In the present study, our aim was to investigate the potential diagnostic value of circRNAs in PCOS.
Methods: The circRNA dataset GSE145296, mRNA dataset GSE155489 and miRNA GSE138572 were downloaded from Gene Expression Omnibus (GEO) database. Then, differentially expressed genes (DEGs) were identified. Based on the potential interactions, a network of cirRNA-related competing endogenous RNAs (ceRNAs) was constructed. Biological functions were predicted by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. For further validation, qRT-PCR method was used to detect the expression level of the candidate circRNAs. Then, receiver operating characteristics (ROC) were constructed to evaluate the diagnostic value of the three differentially expressed circRNA (DE-circRNA).
Results: We constructed a network of cirRNA-related ceRNA network. Hsa_circ_0075691, hsa_circ_0075692 and hsa_circ_0085997 were validate to be dysregulated in PCOS.
Conclusion: Hsa_circ_0075691, hsa_circ_0075692 and hsa_circ_0085997 may be potential diagnostic biomarkers of PCOS, but their specific regulatory mechanisms still need to be further studied.
Keywords: polycystic ovary syndrome, circRNA, biomarkers, bioinformatics
Introduction
PCOS is a common endocrine and metabolic disease in reproductive women, which is characterized by polycystic ovaries, hyperandrogenemia and chronic anovulation.1 However, the health risks of PCOS go well beyond these commonalities, showing symptoms that extend into the reproductive period and even menopause.2 In addition, patients with PCOS are at increased risk of developing metabolic diseases with increasing age, such as obesity,3 cardiovascular disease4 and so on. However, the treatment of PCOS still remains a challenge. Therefore, it is of great significance to study its pathogenesis and search for more effective biomarkers and treatment strategies. Follicular fluid (FF) has proved to provide an important microenvironment for follicular development and oocyte maturation. It is the medium for bidirectional communication between oocytes and the surrounding cells, such as cumulus cells.5 The components of FF are very complex including proteins, RNAs and metabolites,6–10 which are secreted by theca cells, granulosa cells and oocytes.6 Recent studies have shown that circRNAs in FF play an important role in PCOS.11 Huang et al reported that depletion of exosomal circLDLR in follicle fluid derepressed miR-1294 function and inhibits estradiol production via CYP19A1 in PCOS.1 Otherwise, circDDX10 in granulosa cells (GCs) which derived from human follicular fluid may participate in the regulation of ovarian function by affecting the proliferation and apoptosis of GCs and steroid hormone synthesis.12
CircRNA is a kind of RNAs with loop structures produced by abnormal transcript splicing.13 The study of circRNAs has been applied to many important fields spans over 30 years. In recent years, most of the research work on circRNAs has revealed its molecular functions and mechanisms in more detail, and its biological functions include translation, the regulation of gene transcription and miRNA sponge activity.14 Among them, the functions of circRNAs - competing sponge to endogenous RNA to regulate RNA expression and affect RNA function - have attracted considerable scientific attention.15 All these functions suggested that circRNAs may be potential novel diagnostic and prognostic biomarkers of disease.16 So far, it has been reported that circRNAs are extensively involved in the processes of many human diseases, including a variety of cancers,17–19 neurological disorders,20,21 cardiovascular disease,22–24 immune disease25 and so on. Similarly, many circRNAs have been proved to be regulate the process of PCOS, such as circASPH,26 circLDLR1 and circPUM1.27 However, the mechanism of the involvement of much more circRNAs in PCOS remains unclear. Otherwise, the structure of circRNA is stable, not easy to degrade,28 and highly expressed in ovaries,29 which made it to be a proper diagnosis strategy for PCOS.
In the present study, we selected three datasets of PCOS from the GEO datasets, aimed to find novel circRNAs as potential biomarkers for the early diagnosis of PCOS. We also performed a circRNA-miRNA-mRNA regulatory network to identify circRNA that are critical to the biological processes of PCOS. Finally, qRT-PCR was used to validate our bioinformatics analyses. The workflow of our study is shown in Figure 1.
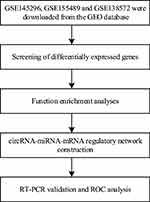 |
Figure 1 The workflow of the study. |
Materials and Methods
Clinical Samples
In our study, a total of 16 PCOS patients and 16 healthy people were collected from Fujian Maternity and Child Health Hospital, Affiliated Hospital of Fujian Medical University. Our study complies with the Declaration of Helsinki. All the research protocol for this study was approved by the Ethics Committee of Fujian Provincial Maternity and Children’s Hospital. PCOS patients were diagnosed according to the revised Rotterdam European Society of Human Reproduction and Embryology/American Society for Reproductive Medicine Criteria (2004). The PCOS patients who fulfilled at least two of the following criteria were included: chronic oligo-ovulation or anovulation, androgen excess and polycystic ovaries. The control group contained patients undergoing IVF or ICSI due to male factors or tubal factors of infertility, who had regular menstrual cycles, normal ovary sonographs and normal ovulating. All of the patients had no history of drugs affecting glucose and lipid metabolism and were without any known medical conditions or diseases, including Cushing’s syndrome, congenital adrenal hyperplasia, androgen-secreting tumors and endometriosis. The follicular fluid was separated from the subscribers. The FF samples were stored in a refrigerator at −80°C for RNA extraction. Informed consent was obtained from all the participants.
Data Sets Collection and Differential Expression Analysis
The RNA expression profiles of PCOS were downloaded from the National Center of Biotechnology Information Gene Expression Omnibus (http: //www.ncbi.nlm.nih.gov/gds/). The GSE14529630 dataset is composed of 12 circRNA expression data of cumulus granulosa cells, including 6 PCOS patients and 6 normal control individuals. For dataset GSE155489, we chose 8 granulosa cells samples for differential mRNA analysis. GSE13857231 dataset contains the microRNA profiles of 10 samples, including 5 PCOS patients and 5 controls. The R software package was used to process the downloaded files, and the unqualified data are converted and removed. After the gene expression matrix was merged, the ensemble IDs were converted to gene names. The data were calibrated, normalized by using the edgeR package in R software, and then log2 transformed. The differentially expressed circRNAs, mRNAs and miRNAs were screened by Limma package, with a threshold of |log2(fold change) [FC]| > 0.58 and P-value < 0.05.
Function Enrichment Analyses
The differentially expressed circRNAs related genes were enriched by GO and KEGG analysis. GO contains three terms: biological processes (BP), cellular components (CC) and molecular functions (MF). The p value < 0.05 indicated significant GO term enrichment among differentially expressed genes. DAVID database (version 6.8; http://david.abcc.ncifcrf.gov) was used for pathway analysis.32 P value < 0.05 was considered as statistically significant.
ceRNA Network Construction
The circular RNA Interactome (CircInteractome) (https://circinteractome.nia.nih.gov/) database was used to predict the miRNA-binding sites (MREs). Next, we used miRTarBase33 (http://mirtarbase.mbc.nctu.edu.tw) and TargetScan34 (http://www.targetscan.org) databases to predict the interactions between miRNAs and mRNAs. Only the mRNAs recognized by both databases were considered as candidate mRNAs, and were intersected with DE-mRNAs to screen the DE-mRNAs targeted by DE-miRNAs. The circRNA-miRNA-mRNA regulatory network was constructed using a combination of circRNA-miRNA pairs and miRNA-mRNA pairs. Finally, the network was visualized and mapped using Cytoscape v3.6.1.
Validation with qRT- PCR Method
The potential expression of circRNAs was confirmed by qRT-PCR. Total RNA was extracted from the follicular fluid using TRIzol Reagent (Invitrogen) according to the manufacturer’s protocol. RNA purity was measured using a NanoDrop Spectrophotometer (Nanodrop, Thermo Fisher Scientific). The total PCR reaction of circRNA was 20μL, including 6 μL of cDNA, 10 μL of SYBR Green, and 0.5 μL of primers for forward (10 μmol/L) and reverse (10 μmol/L). GAPDH as an internal reference. Primers sequences are summarized in Table 1. All PCR reactions were conducted in triplicate. The relative expression of circRNAs was calculated by 2−ΔΔCt method.
 |
Table 1 Specific RNAs Primers for qRT-PCR Analysis |
Statistical Analysis
Data were analyzed using GraphPad Prism 8.0 (GraphPad software, San Diego, USA). The data are expressed as means ± SD. Unpaired t-test was used to distinguish the differences between the two groups. P value < 0.05 was considered statistically significant with 2-tailed tests. Receiver operating characteristic (ROC) curve was used to analyze the diagnostic value, including the area under the curve (AUC), sensitivity, and specificity.
Results
Identification of Differentially Expressed Genes
Three datasets GSE145296, GSE155489 and GSE138572 were downloaded from the GEO database, respectively. The volcano and heatmap showed differential expression profiles of the PCOS and healthy samples (Figure 1). A total of 1591 DE-circRNAs were screened from GSE145296 dataset, including 945 upregulated and 646 downregulated circRNAs (Figure 2A and D). The DE-mRNAs and DE-miRNAs of PCOS patients and healthy controls, obtained from datasets GSE155489 and GSE138572, respectively. We totally identified 1688 DE-mRNAs (739 upregulated and 949 downregulated) (Figure 2B and E) and 38 DE-miRNAs (18 upregulated and 20 downregulated) (Figure 2C and F).
Functional Enrichment Analysis of DE-circRNAs in PCOS
A total of 1591 DE-circRNAs were identified. Among these genes, hsa_circ_0017962, hsa_circ_0056003 and hsa_circ_0032879 were most significant. In order to investigate the potential function of the DE-circRNAs. GO and KEGG enrichment analysis was used to explore the functions of DE-circRNAs host genes. The top ten most significant enrichment BP, CC and MF terms are shown in Figure 3A–C. From our results, the top 3 enriched BP terms are response to oxygen levels, response to decreased oxygen levels and response to hypoxia. This may indicate that PCOS is in an anoxic environment. The top three enriched CC terms included collagen-containing extracellular matrix, mitochondrial protein complex, and mitochondrial matrix. The top three enriched MF terms included extracellular matrix structural constituent, peptide antigen binding and collagen binding. Otherwise, a total of 52 pathways were enriched. The top ten pathways were shown in Figure 3D. Among them, we found metabolism-related pathways were significantly enriched including Valine, leucine and isoleucine degradation, Propanoate metabolism, Pyruvate metabolism, Carbon metabolism, Glyoxylate and dicarboxylate metabolism, Citrate cycle (TCA cycle), which may suggest an active metabolism in PCOS.
A circRNA-miRNA-mRNA Regulatory Network of PCOS
Subsequently, we constructed a circRNA-miRNA-mRNA regulatory network to reveal the connections of circRNA, miRNA and mRNA in PCOS (Figure 4), which composed with 16 nodes and 14 edges. As shown in Figure 4, there were only three circRNAs (hsa_circ_0075691, hsa_circ_0075692 and hsa_circ_0085997) and three miRNAs (hsa-miR-320c, hsa-miR-455-5p and hsa-miR-346) in the network. The basic characteristics of the 3 circRNAs are listed in Table 2. Their basic structural pattern is shown in Figure 5.
 |
Table 2 Basic Characteristics of the Candidate circRNAs |
 |
Figure 5 The basic structural patterns of the three DE-circRNAs. Structural patterns of the 3 DE-circRNAs by circRNADb (http://reprod.njmu.edu.cn/circrnadb/circRNADb.php). (A) Structure of hsa_circ_0075691. (B) Structure of hsa_circ_0075692. (C) None. |
Validation the Expression of the circRNAs
To further evaluate the expression of the three circRNAs, a total of 16 PCOS patients and 16 healthy controls were collected to validate the analyses results by qRT-PCR. Consistent with the omics data, hsa_circ_0075691 and hsa_circ_0075692 were significantly upregulated (Figure 6A and B), hsa_circ_0085997 was downregulated (Figure 6C) between PCOS and controls. To assess the potential diagnosis value of the three circRNAs for PCOS, we further implemented a ROC curve analysis. We found that ROC curve of hsa_circ_0075691 showed a significant distinguishing efficiency with an AUC value of 0.89 (95% CI: 0.65–0.96, **p <0.01) (Figure 6D), an AUC value of 0.80 for hsa_circ_0075692 (95% CI: 0.78–1.00, **p <0.01) (Figure 6E) and an AUC value of 0.75 for hsa_circ_0085997 (95% CI: 0.57–0.93, **p <0.01) (Figure 6F). These indicated that circRNAs could be candidate biomarkers for PCOS diagnosis.
Discussion
PCOS, with its high incidence among women of reproductive age and multiple side effects, has become a global health burden and brings unpleasant life experiences to patients.35 The etiology of PCOS is complex with many genetic mutations, so the clinical management is faced with difficulties.36 Therefore, it is urgent to find effective and efficient targets for the treatment of PCOS. With the development of high throughput sequencing, studies applying this technique for assessing PCOS have revealed the differential expression of transcripts in PCOS. Recently, the expression profile of circRNAs in polycystic ovary syndrome has been partially reported.29,37 Circular RNAs are spliced endogenous RNAs that can potentially be used as diagnostic and prognostic biomarkers for disease.38–40 Nevertheless, there were only a few studies have reported the diagnostic value of circRNAs in PCOS. Recent studies showed that circASPH26 and hsa_circ_011853041 may have the potential to be used as diagnostic biomarkers of PCOS. These findings give us a direction for our research. In this study, the differentially expressed circRNAs hsa_circ_0075691, hsa_circ_0075692 and hsa_circ_0085997 have not been previously reported in other diseases, suggesting that the specificity and effectiveness of these circRNA could be potential biomarkers of PCOS.
As we know, circRNAs have a large number of binding sites to sponge miRNAs, which could competitively bind to the miRNAs, reducing the inhibition of target genes. In the study, we used three different datasets to construct a circRNA-miRNA-mRNA network in PCOS. Studying the function of these DE-circRNAs is a very interesting project. Through the GO analysis, we found that the biological process Response to hypoxia was significantly enriched. Otherwise, Wang and Wu has reported that hypoxia inducible factor-1 (HIF-1) decreased in granulosa cells of polycystic ovary syndrome, while the levels of reactive oxygen species increased.42 Similarly, Zhao et al found the protein and mRNA level of HIF-1α was significantly downregulated of PCOS endometrial tissues than that in healthy control.43 These may indicated that the HIF pathway is inhibited in the ovary of PCOS and that PCOS causes the ovary to be in a peroxidation state. In addition, pathway analysis showed many metabolism-related pathways were significantly enriched, such as Valine, leucine and isoleucine degradation, Propanoate metabolism, Pyruvate metabolism, Carbon metabolism, Glyoxylate and dicarboxylate metabolism. As we know, PCOS is one of the most common endocrine diseases in women, so endocrine disruption obviously causes the changes in various metabolic pathways. And our findings are consistent with previous reports.44–46
CeRNA network reveals a new mechanism of RNA interaction, greatly expanding the functional genetic information in the human genome and playing important roles in pathological conditions.47 In the present study, we aimed to constructed a ceRNA network to reveal potential RNAs regulation relationship in PCOS. Through our bioinformatics analyses, three circRNAs was met to the ceRNA network. To validate the expression of the three genes, more samples were used to make a validation set. ROC analysis showed good specificity of the candidate genes. CircRNA-miRNA-mRNA network implied that circRNA may function by sponging miRNAs. MiR-346 has been reported to be upregulated under hypoxia in renal cell carcinoma.48 Otherwise, miR-346 was upregulated in primary sclerosing cholangitis and associated with TNF-α signaling pathway.49 So, we speculate that hsa_circ_0085997 may absorb miR-346 to involved in inflammatory regulation in PCOS. However, the function of hsa-mir-320c and hsa-mir-455-5p were not reported in detail. The functions and accurate regulatory mechanisms of the three circRNAs were need more validation. In a word, integrated bioinformatics and validation, we found three potential circRNA biomarkers to diagnose PCOS, but there remains further study of the complex regulatory mechanisms in PCOS.
Conclusion
In conclusion, through integrated bioinformatics, a circRNA related ceRNA network was constructed. Three circRNAs (hsa_circ_0075691, hsa_circ_0075692 and hsa_circ_0085997) were associated with the diagnosis and treatment of PCOS by RT-PCR and ROC curve analysis. However, there are still some limits about the molecular function and regulatory mechanism of the circRNAs in this study. We will focus on this in the following work.
Data Sharing Statement
All datasets generated for this study are included in the article materials.
Acknowledgments
This work was supported by General Project of Fujian Natural Science Foundation (2019J01052054), Key project of Fujian Provincial Maternity and Child Health Hospital Science and Technology Innovation Startup Fund Department (YCXZ18-04), Health research project of Department of Finance ([2020] No. 467) and Fujian Natural Science Foundation (2019J01511).
Disclosure
The authors declare that they have no competing interests.
References
1. Huang X, Wu B, Chen M, et al. Depletion of exosomal circLDLR in follicle fluid derepresses miR-1294 function and inhibits estradiol production via CYP19A1 in polycystic ovary syndrome. Aging. 2020;12(15):15414–15435. doi:10.18632/aging.103602
2. Christakou C, Diamanti-Kandarakis E. Polycystic ovary syndrome–phenotypes and diagnosis. Scand J Clin Lab Invest Suppl. 2014;244:
3. Kandaraki E, Christakou C, Diamanti-Kandarakis E. Metabolic syndrome and polycystic ovary syndrome … and vice versa. Arq Bras Endocrinol Metabol. 2009;53(2):227–237. doi:10.1590/S0004-27302009000200014
4. Carmina E, Bucchieri S, Mansueto P, Rini G, Ferin M, Lobo RA. Circulating levels of adipose products and differences in fat distribution in the ovulatory and anovulatory phenotypes of polycystic ovary syndrome. Fertil Steril. 2009;91(4 Suppl):1332–1335. doi:10.1016/j.fertnstert.2008.03.007
5. Matzuk MM, Burns KH, Viveiros MM, Eppig JJ. Intercellular communication in the mammalian ovary: oocytes carry the conversation. Science. 2002;296(5576):2178–2180. doi:10.1126/science.1071965
6. Rodgers RJ, Irving-Rodgers HF. Formation of the ovarian follicular antrum and follicular fluid. Biol Reprod. 2010;82(6):1021–1029. doi:10.1095/biolreprod.109.082941
7. Ambekar AS, Nirujogi RS, Srikanth SM, et al. Proteomic analysis of human follicular fluid: a new perspective towards understanding folliculogenesis. J Proteomics. 2013;87:68–77. doi:10.1016/j.jprot.2013.05.017
8. Tu M, Wu Y, Mu L, Zhang D. Long non-coding RNAs: novel players in the pathogenesis of polycystic ovary syndrome. Ann Transl Med. 2021;9(2):173. doi:10.21037/atm-20-5044
9. Lefevre PLC, Nardelli TC, Son WY, et al. Polybrominated diphenyl ethers in human follicular fluid dysregulate mural and cumulus granulosa cell gene expression. Endocrinology. 2021;162(3). doi:10.1210/endocr/bqab003
10. Zhao H, Wang L, Wang Y. Circulating microRNAs as candidate biomarkers for the ovarian response during in vitro fertilization. Medicine. 2021;100(6):e24612. doi:10.1097/MD.0000000000024612
11. Wang LP, Peng XY, Lv XQ, et al. High throughput circRNAs sequencing profile of follicle fluid exosomes of polycystic ovary syndrome patients. J Cell Physiol. 2019;234(9):15537–15547.
12. Cai H, Chang T, Li Y, et al. Circular DDX10 is associated with ovarian function and assisted reproductive technology outcomes through modulating the proliferation and steroidogenesis of granulosa cells. Aging. 2021;13(7):9592–9612. doi:10.18632/aging.202699
13. Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19(1):172. doi:10.1186/s12943-020-01286-3
14. Verduci L, Tarcitano E, Strano S, Yarden Y, Blandino G. CircRNAs: role in human diseases and potential use as biomarkers. Cell Death Dis. 2021;12(5):468. doi:10.1038/s41419-021-03743-3
15. Huang S, Yang B, Chen BJ, et al. The emerging role of circular RNAs in transcriptome regulation. Genomics. 2017;109(5–6):401–407. doi:10.1016/j.ygeno.2017.06.005
16. Sheng JQ, Liu L, Wang MR, Li PY. Circular RNAs in digestive system cancer: potential biomarkers and therapeutic targets. Am J Cancer Res. 2018;8(7):1142–1156.
17. Wang C, Jiang Y, Lei Q, et al. Potential diagnostic and prognostic biomarkers of circular RNAs for lung cancer in China. Biomed Res Int. 2019;2019:8023541.
18. Taborda MI, Ramirez S, Bernal G. Circular RNAs in colorectal cancer: possible roles in regulation of cancer cells. World J Gastrointest Oncol. 2017;9(2):62–69. doi:10.4251/wjgo.v9.i2.62
19. Wang S, Zhang K, Tan S, et al. Circular RNAs in body fluids as cancer biomarkers: the new frontier of liquid biopsies. Mol Cancer. 2021;20(1):13. doi:10.1186/s12943-020-01298-z
20. Xie L, Mao M, Xiong K, Jiang B. Circular RNAs: a novel player in development and disease of the central nervous system. Front Cell Neurosci. 2017;11:354. doi:10.3389/fncel.2017.00354
21. Li L, Ni Z, Si X, et al. Emerging clues of regulatory roles of circular RNAs through modulating oxidative stress: focus on neurological and vascular diseases. Oxid Med Cell Longev. 2021;2021:6659908.
22. Ward Z, Pearson J, Schmeier S, Cameron V, Pilbrow A. Insights into circular RNAs: their biogenesis, detection, and emerging role in cardiovascular disease. RNA Biol. 2021;1–18. doi:10.1080/15476286.2021.1891393
23. Kishore R, Garikipati VNS, Gonzalez C. Role of circular RNAs in cardiovascular disease. J Cardiovasc Pharmacol. 2020;76(2):128–137. doi:10.1097/FJC.0000000000000841
24. Zhang L, Zhang Y, Wang Y, Zhao Y, Ding H, Li P. Circular RNAs: functions and clinical significance in cardiovascular disease. Front Cell Dev Biol. 2020;8:584051. doi:10.3389/fcell.2020.584051
25. Liu H, Zou Y, Chen C, Tang Y, Guo J. Current understanding of circular RNAs in systemic lupus erythematosus. Front Immunol. 2021;12:628872. doi:10.3389/fimmu.2021.628872
26. Wu G, Xia J, Yang Z, et al. CircASPH promotes KGN cells proliferation through miR-375/MAP2K6 axis in polycystic ovary syndrome. J Cell Mol Med. 2020. doi:10.1111/jcmm.16231
27. Deng L, Chen Q, Xie J, Wei W, Hui H. circPUM1 promotes polycystic ovary syndrome progression by sponging to miR-760. Gene. 2020;754:144903. doi:10.1016/j.gene.2020.144903
28. Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol. 2017;14(8):1035–1045. doi:10.1080/15476286.2016.1271524
29. Che Q, Liu M, Xu J, et al. Characterization of circular RNA expression profiles in cumulus cells from patients with polycystic ovary syndrome. Fertil Steril. 2019;111(6):1243–1251 e1241. doi:10.1016/j.fertnstert.2019.02.023
30. Ma Z, Zhao H, Zhang Y, Liu X, Hao C. Novel circular RNA expression in the cumulus cells of patients with polycystic ovary syndrome. Arch Gynecol Obstet. 2019;299(6):1715–1725. doi:10.1007/s00404-019-05122-y
31. Mao Z, Li T, Zhao H, Qin Y, Wang X, Kang Y. Identification of epigenetic interactions between microRNA and DNA methylation associated with polycystic ovarian syndrome. J Hum Genet. 2021;66(2):123–137. doi:10.1038/s10038-020-0819-6
32. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi:10.1038/nprot.2008.211
33. Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43(Databaseissue):D146–152. doi:10.1093/nar/gku1104
34. Fromm B, Billipp T, Peck LE, et al. A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome. Annu Rev Genet. 2015;49:213–242. doi:10.1146/annurev-genet-120213-092023
35. Costello MF, Misso ML, Balen A, et al. Evidence summaries and recommendations from the international evidence-based guideline for the assessment and management of polycystic ovary syndrome: assessment and treatment of infertility. Hum Reprod Open. 2019;2019(1):hoy021. doi:10.1093/hropen/hoy021
36. Gibson-Helm M, Teede H, Dunaif A, Dokras A. Delayed diagnosis and a lack of information associated with dissatisfaction in women with polycystic ovary syndrome. J Clin Endocrinol Metab. 2017;102(2):604–612.
37. Zhang C, Liu J, Lai M, et al. Circular RNA expression profiling of granulosa cells in women of reproductive age with polycystic ovary syndrome. Arch Gynecol Obstet. 2019;300(2):431–440. doi:10.1007/s00404-019-05129-5
38. Xu Y, Xu X, Ocansey DKW, et al. CircRNAs as promising biomarkers of inflammatory bowel disease and its associated-colorectal cancer. Am J Transl Res. 2021;13(3):1580–1593.
39. Yang T, Li Y, Zhao F, Zhou L, Jia R. Circular RNA Foxo3: a promising cancer-associated biomarker. Front Genet. 2021;12:652995. doi:10.3389/fgene.2021.652995
40. Liu Y, Li J, Bu H, et al. Circular RNA expression alteration identifies a novel circulating biomarker in serum exosomal for detection of alcohol dependence. Addict Biol. 2021;e13031. doi:10.1111/adb.13031
41. Jia C, Wang S, Yin C, Liu L, Zhou L, Ma Y. Loss of hsa_circ_0118530 inhibits human granulosa-like tumor cell line KGN cell injury by sponging miR-136. Gene. 2020;744:144591. doi:10.1016/j.gene.2020.144591
42. Wang J, Wu X. The effects of mitochondrial dysfunction on energy metabolism switch by HIF-1alpha signalling in granulosa cells of polycystic ovary syndrome. Endokrynol Pol. 2020;71(2):134–145. doi:10.5603/EP.a2020.0002
43. Zhao D, Qu Q, Dai H, et al. Effects of hypoxia-inducible factor‑1α on endometrial receptivity of women with polycystic ovary syndrome. Mol Med Rep. 2018;17(1):414–421.
44. Zheng R, Qing P, Han M, et al. The effect of acupuncture on glucose metabolism and lipid profiles in patients with PCOS: a systematic review and meta-analysis of randomized controlled trials. Evid Based Complement Alternat Med. 2021;2021:5555028. doi:10.1155/2021/5555028
45. Neubronner SA, Indran IR, Chan YH, Thu AWP, Yong EL. Effect of Body Mass Index (BMI) on phenotypic features of Polycystic Ovary Syndrome (PCOS) in Singapore women: a prospective cross-sectional study. BMC Womens Health. 2021;21(1):135. doi:10.1186/s12905-021-01277-6
46. Burbach BJ, O’Flanagan SD, Shao Q, et al. Irreversible electroporation augments checkpoint immunotherapy in prostate cancer and promotes tumor antigen-specific tissue-resident memory CD8+ T cells. Nat Commun. 2021;12(1):3862. doi:10.1038/s41467-021-24132-6
47. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
48. Su ZH, Liao HH, Lu KE, et al. Hypoxia-responsive miR-346 promotes proliferation, migration, and invasion of renal cell carcinoma cells via targeting NDRG2. Neoplasma. 2020;67(5):1002–1011. doi:10.4149/neo_2020_190917N915
49. Kempinska-Podhorodecka A, Blatkiewicz M, Wunsch E, et al. Oncomir MicroRNA-346 is upregulated in colons of patients with primary sclerosing cholangitis. Clin Transl Gastroenterol. 2020;11(1):e00112. doi:10.14309/ctg.0000000000000112
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.