Back to Journals » Neuropsychiatric Disease and Treatment » Volume 15
Identification of key genes, pathways and miRNA/mRNA regulatory networks of CUMS-induced depression in nucleus accumbens by integrated bioinformatics analysis
Authors Ma K , Zhang HX, Wei GH, Dong ZF, Zhao HJ, Han XC, Song XB, Zhang HL, Zong X, Baloch Z , Wang SJ
Received 3 January 2019
Accepted for publication 10 February 2019
Published 14 March 2019 Volume 2019:15 Pages 685—700
DOI https://doi.org/10.2147/NDT.S200264
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yuping Ning
Ke Ma,1 Hongxiu Zhang,2 Guohui Wei,1 Zhenfei Dong,1 Haijun Zhao,1 Xiaochun Han,1 Xiaobin Song,1 Huiling Zhang,1 Xin Zong,1 Zulqarnain Baloch,3 Shijun Wang1
1Shandong Co-Innovation Center of Classic TCM formula, Shandong University of Traditional Chinese Medicine, Jinan, Shandong 250355, People’s Republic of China; 2Institute of Virology, Jinan Center for Disease Control and Prevention, Jinan 250021, People’s Republic of China; 3College of Veterinary Medicine, South China Agricultural University, Guangzhou 510642, People’s Republic of China
Introduction: Major depressive disorder (MDD) is a recurrent, devastating mental disorder, which affects >350 million people worldwide, and exerts substantial public health and financial costs to society. Thus, there is a significant need to discover innovative therapeutics to treat depression efficiently. Stress-induced dysfunction in the subtype of neuronal cells and the change of synaptic plasticity and structural plasticity of nucleus accumbens (NAc) are implicated in depression symptomology. However, the molecular and epigenetic mechanisms and stresses to the NAc pathological changes in depression remain elusive.
Materials and methods: In this study, treatment group mice were treated continually with the chronic unpredictable mild stress (CUMS) until expression of depression-like behaviors were found. Depression was confirmed with sucrose preference, novelty-suppressed feeding, forced swimming, and tail suspension tests. We applied high-throughput RNA sequencing to assess microRNA expression and transcriptional profiles in the NAc tissue from depression-like behaviors mice and control mice. The regulatory network of miRNAs/mRNAs was constructed based on the high-throughput RNA sequence and bioinformatics software predictions.
Results: A total of 17 miRNAs and 10 mRNAs were significantly upregulated in the NAc of CUMS-induced mice with depression-like behaviors, and 12 miRNAs and 29 mRNAs were downregulated. A series of bioinformatics analyses showed that these altered miRNAs predicted target mRNA and differentially expressed mRNAs were significantly enriched in the MAPK signaling pathway, GABAergic synapse, dopaminergic synapse, cytokine–cytokine receptor interaction, axon guidance, regulation of autophagy, and so on. Furthermore, dual luciferase report assay and qRT-PCR results validated the miRNA/mRNA regulatory network.
Conclusion: The deteriorations of GABAergic synapses, dopaminergic synapses, neurotransmitter synthesis, as well as autophagy-associated apoptotic pathway are associated with the molecular pathological mechanism of CUMS-induced depression.
Keywords: stress, depression, nucleus accumbens, miRNA, mRNA
Introduction
Major depressive disorder (MDD) is a common, diverse, widespread, and intermittent neuropsychiatric burden that includes the wide range of symptoms, such as inability to feel pleasure, lack of motivation, retraction from social interaction, cognitive difficulties, and changes in appetite.1,2 Depression affects a significant portion of the world’s population annually; according to the World Health Organization, it is the leading cause of disability in the world.3 Extensive research efforts have been done in past decades; however, the etiology of depression is still indefinable, its diagnosis unclear, and the pharmacotherapy ineffective. This may be due to poor understanding of the molecular pathophysiology of depression.4
It has been established that depression is a collection of diseases, with overlapping causal pathways, beginning with interaction among genetic and environmental factors.5,6 The persistent stress to the genetically susceptible persons directs to the deficits of hypothalamus–pituitary–adrenal axis, neurotrophic factors, monoamine factor, and cytokine, which induce neuron atrophy and dysfunction in brain reward circuits, including the nucleus accumbens (NAc), prefrontal cortex (PFC), ventral tegmental area (VTA), and basolateral amygdala, in the depression patients and animal models with depression-like behaviors.7,8 A number of abnormally regulated genes have been reported in genetically susceptible persons.9–11 However, existing data cannot explain mechanism that mediates pathological changes in depression. Thus, it is important to investigate the potential changes in gene expressions and the regulation of these changes, specifically in brain regions that mediate emotional memory and anxiety.
It has been suggested that NAc is essential for emotion processing and receives dense innervations from mood-related structures including PFC, VTA, hippocampus, amygdala, and hypothalamus.12,13 It is regarded as a neural interface between action and motivation, and has an important role in food intake, sexual behavior, reward-motivated behavior, and stress-related behavior.14 The dysfunction in this region may be correlated to low motivation, interest loss, and anhedonia in depression disorder. Recently, various reports have suggested that synaptic plasticity, structural plasticity changes in NAc medium spiny neurons, and reduced activity are implicated in depression symptomology.15–18 Recently, neuromodulation, an intervention targeting the NAc, has been used in patients diagnosed with treatment-resistant depression.19,20 Furthermore, NAc deep brain motivation has also been linked with anxiolytic and antidepressant effect and enhancement of quality of life in patients diagnosed with severe treatment-resistant depression.21
Gene expression disturbance in the brain is most probably linked with environmental factors which lead to depression.22,23 This assumption has been established by analyzing miRNA in depressive patients24,25 and in mice with depression-like behaviors.26 A reaction chain from stress to miRNA dysregulation, alternations in mRNA/protein expression, and neuron atrophy and function deficit are proposed to be associated with MDD. However, the molecules acknowledged for major depression are exhibited differently in different reports. The inconsistency may be due to different experimental models in different studies.
This study was designed to characterize the NAc transcriptional and post-transcriptional landscape in mice with depression-like behaviors encouraged by chronic unpredictable mild stress (CUMS). We performed an unbiased, genome-wide bioinformatics analysis of mRNA transcriptome profiles and miRNA in the NAc from CUMS-induced depression mice with the use of high-throughput RNA sequencing. The miRNA–mRNA networks were created to probe and illustrate the complexity of stress-induced miRNA regulation. By using different associated analyses and comparisons, we found signal pathways in the NAc associated with stress-induced depression, in order to present the guidelines for understanding the molecular mechanisms of depression and exploring novel therapeutic strategies for this complex disorder.
Materials and methods
Animals
Male C57BL/6J mice (4 weeks, 16–18 g) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd (Beijing, China). In this study, all procedures were carried out under the Guideline of National Institutes of Health, USA, for the Care and Use of Laboratory Animals and approved by Animal Care and Use Committee of Shandong University of Traditional Chinese Medicine Institutional (SDUTCM201805311223).
CUMS paradigm
The CUMS paradigms were performed with a minor modification according to the previous studies.7,27 All experimental mice were acclimated to environment and sucrose intake for 1 week. We divided the experimental mice into two groups: control and CUMS. The 3–4 mice in the control groups were housed in one cage, and each mouse in the CUMS group was housed in a single cage. The control group was kept in undisturbed cages. A variate-stressor paradigm was used in the CUMS group.
The CUMS protocol consisted of 13 stressors including social isolation, food and water deprivation, empty bottles, soiled cage, restraint space, circadian disturbance, tilted cage, stroboscope, cold (4°C), rotating cage, wet cage, white noise, and space reduction (Table S1). This experiment was maintained for 4 weeks until the mice expressed depression-like behavior phenotype.
Behavioral assessments
Behavioral tests were conducted to assess whether the CUMS induced mice expressed depressed phenotype. The novelty-suppressed feeding test (NST) and sucrose preference test (SPT) were used to assess the anhedonia, and the tail suspension test (TST) and forced swimming test (FST) were used to estimate the behavioral despair. Behavioral assessment experiments were performed in the light cycle with a sound-proof behavioral facility.
TST and FST
TST and FST were carried out as previously reported.28
SPT
SPT was conducted with a minor modification according to previous studies.29,30
NST
Mice were observed for NST after 24-hour food deprivation (water was provided ad libitum). Details of the experiment have been previously reported.30
RNA purification and sequencing
The protocol of RNA purification and sequencing has already been reported in our previously published study.7 In brief, if there were depression-like behaviors 24 hours after treatment (CUMS), both control and CUMS mice were anesthetized by using isoflurane and decapitated by guillotine. NAc samples were dissected and immediately placed on the ice-cold glass slides. These tissues were placed into the frozen vials that contained RNA and stored at −80°C for subsequent analyses. Total RNA was isolated from the colon with the RNAiso Plus Kit (TaKaRa, Dalian, China). After extracting RNA, the UV absorbance was used to measure RNA purity and quality. Absorbance at 260 nm was used to measure the amount of nucleic acid present in the sample. Absorbance at 280 nm were used to estimate the amount of protein in the sample. Absorbance at 230 nm was used to determine the amount of other contaminants that may be present in the samples, such as guanidine thiocyanate. The A260/A280 ratios ranging from 1.9 to 2.1 were acceptable ratios for purity in this experiment, and the requirement for A260/A230 ratios was >2.0. RNA concentration was calculated using the 260 nm reading. The samples with total RNA amount >10 μg, the concentration >200 ng/μL, were selected for the construction of transcriptome and small RNA libraries, respectively. The sequencings were done with the use of BGISEQ-500 platform (BGI, Shenzhen, China). The average reading length of two libraries was about 100 bp (pair-end) and 50 bp (single-end).
Bioinformatics
Bioinformatics analysis of transcriptome, miRNA expression profile, and integrated miRNA/mRNA network have been reported in our previous report.7,30
Quantitative RT-PCR for the validations of miRNA and mRNA
To validate the RNA sequencing data, qRT-PCR was used to detect five miRNAs and five mRNAs which were significantly different between control and mice with depression-like behaviors (n=12/group), in which the tissues were included as those samples for sequencing (all gene primers are listed in Table S2). Briefly, total RNA was harvested from the NAc with the RNAiso Plus Kit (TaKaRa). Then, RNA was reverse-transcribed into cDNA with PrimeScript™ RT reagent Kit (TaKaRa). qRT-PCR was carried out using SYBR® Premix Ex Taq™ kit (TaKaRa) on biosystems QuantStudio 7 Flex (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. The relative expression level of mRNAs in the NAc was normalized to GAPDH. The relative miRNA expression in the NAc was normalized to U6. The 2−ΔΔCt method was used to calculate the qRT-PCR results. Each sample of qRT-PCR was repeated in three replications.
Dual luciferase reporter assay
The 3′-untranslated region (UTR) sequence of Gad1 mRNA was amplified and fused into the NotI and XhoI sites of psiCHECK2. The site-directed mutation of the detected miR-144-3p targeting site of Gad1 mRNA 3′-UTR was constructed with the guideline of Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). The miRNA mimics and the negative control (primer sequences are listed in Table S3) were synthesized in Guangzhou Rui Bo Biological Technology Co., Ltd. HEK 293 T cells (24-well plates) were transfected with reporter vectors together with miR-144-3p mimics and inhibitor or the corresponding miR-NC. The renilla luciferase activity was measured and normalized to firefly luciferase activity at 48 hours post-transfection by using Dual-Glo® Luciferase Assay System (E2920; Promega Corporation, Fitchburg, WI, USA). Each experiment was performed in triplicate.
Statistical analyses
The data of the behavior tests, gene analyses, and luciferase activity are presented as mean ± SEM. The association between miRNAs and prediction was investigated by Pearson’s correlation coefficients. Unpaired Student’s t-test was used to evaluate the statistical difference between the two groups. P<0.05 was considered statistically significant.
Results
CUMS induces depression-like behaviors
The mood states of CUMS-treated or control group mice were tested by NST, SPT, TST, and FST. Mice exposed to CUMS showed extensively improved immobility in FST (Figure 1A) and TST (Figure 1B) compared to control group, which demonstrated that those mice were in the status of helplessness and hopelessness. Stressed mice also displayed significantly reduced sucrose preference (Figure 1C) and increased latency to feed in the NST (Figure 1D), which showed that those mice were in the mood states of anhedonia and anxiety. Our data indicated that the CUMS paradigm induced depression-like behaviors.
Overall qualities of RNA-sequencing data set
In this study, we used RNA-sequencing and assessed miRNA expression and transcriptional profiles in NAc sample from mice with depression-like behaviors and control mice (n=4 per group). About 32,489,852–35,179,878 raw sequence reads were produced in the small RNA library. After trimming and filtering of reads with low quality and adaptor, a total of 29,759,815–32,525,311 clean small RNA reads were obtained (Table S4). The high qualities of small RNA sequencing and transcriptome data were further used for analysis. In parallel, RNAs from 45,123,294 to 45,175,652 raw sequence reads of about 100 bp were obtained from eight mRNA library Illumina sequencing. After filtering the reads that contained N with adaptor sequence and low quality, about 1.2 GB clean reads from each library were generated and mapped, which were equivalently about 75.82%–80.62% of the total reads from the University of California Santa Cruz mm10 (Table S5). In conclusion, these results from the high qualities of small RNA sequencing and transcriptome data can be used for further analysis.
miRNA expression profiles in NAc from CUMS-induced depression mice
The miRNA expression profile has been given in Table 1 with their expression change >1.5 fold and P-value <0.05. Seventeen miRNAs were significantly upregulated in the NAc of mice with CUMS-induced depression-like behaviors and 12 miRNAs were downregulated. These upregulated miRNAs included certain known miRNAs (mmu-miR-378d, mmu-miR-34b-5p, mmu-miR-34c-3p, mmu-miR-194-5p, mmu-miR-206-3p, mmu-miR-10a-5p, mmu-miR-144-3p, mmu-let-7a-1-3p, mmu-miR-34c-5p, mmu-miR-34b-3p, mmu-miR-21a-5p, mmu-miR-10b-5p, mmu-miR-31-5p, mmu-miR-219b-5p, and mmu-miR-338-3p), as well as two novel miRNAs (novel_mir42 and novel_mir39). Whereas, the downregulated miRNAs included mmu-miR-7a-2-3p, mmu-miR-124-5p, mmu-miR-351-5p, mmu-miR-127-5p, mmu-miR-671-5p, mmu-miR-335-3p, mmu-miR-377-3p, mmu-miR-574-5p, mmu-miR-200b-5p mmu-miR-486a-5p, mmu-miR-532-3p, and novel_mir29.
  | Table 1 miRNAs with quantitative change over 1.5-folds and their characteristics |
In order to confirm the results of miRNA sequencing analysis, three upregulated miRNAs (mmu-miR-10a-5p, mmu-miR-144-3p, and mmu-miR-219b-5p) and two downregulated miRNAs (mmu-miR-124-5p and mmu-miR-127-5p) were selected for performing qRT-PCR. Consistent with high-throughput sequencing, these miRNAs from mice with CUMS-induced depression-like behaviors are significantly changed in qRT-PCR compared with those from control mice (Figure 2A). These consistent results from the miRNA sequencing (Figure 2B) and qRT-PCR supported the validation of our study.
Transcriptional profiles in the NAc from mice with CUMS-induced depression behaviors
mRNAs in the NAc from mice with depression-like behaviors and controls were quantified by sequencing total RNAs. We computed Reads Per Kilobase Million (RPKM) values for those genes. Genes with lower expression level (RPKM <5) were removed, thus 16,918 mRNAs were left for differential expression. The mRNA expression profile has been given in Table 1 in which alterations in their expressions were >1.5-fold in mice with depression-like behaviors and controls with P-value <0.05, which was a criterion to ensure differentially expressed genes. Twenty-nine mRNAs were significantly downregulated in the NAc from mice with CUMS-induced depression-like behaviors and 10 mRNAs were upregulated (Table 2). The decreased expression of mRNAs in the NAc of mice with CUMS-induced depression-like behaviors included Bdnf, Gad1, Atg13, Slc32a1 (VGAT), Gad2, Th, Mbp, and so on. Whereas, compared to control mice, Aspa, Atg2b, Hba-a1, Hba-a2, Hbb-b2, and Tnfsf12 mRNAs were significantly increased in the NAc of mice with CUMS-induced depression-like behaviors.
  | Table 2 mRNAs with quantitative change over 1.5-folds and their characteristics |
Based on the bioinformatics analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) database, these differentially expressed genes (DEGs) were significantly enriched in the following signaling pathways and processes, such as the MAPK signaling pathway, GABAergic synapse, dopaminergic synapse, neuroactive ligand–receptor interaction, glutamate metabolism, cytokine–cytokine receptor interaction, axon guidance, regulation of autophagy, apoptosis, and so on (Table 3). Thus, the GABAergic synapses, dopaminergic synapses, axon guidance, neurotrophin signaling pathway, neurotransmitter synthesis, autophagy-associated apoptotic pathway, and neural-immune process in the NAc were associated with the pathogenesis of depression. It was noteworthy that the unregulated genes were clustered in hemoglobin genes (Hba-a1, Hba-a2, and Hbb-b2).
In order to verify the differentially expressed genes data, five significantly changed mRNAs involved in different cellular functions were selected for performing qRT-PCR from the NAc tissues that had been used for the mRNA sequencing. The level of Hbb-b2 (P<0.001) gene expression was increased, and the expressions of Bdnf, Slc32a1(VGAT), Mbp, and Gad1 were decreased in mice with CUMS-induced depression-like behaviors compared with that in the control mice (all P<0.001, Figure 3A). The consistent results obtained by gene sequencing (Figure 3B) and qRT-PCR supported our research validation.
Integrated miRNA/mRNA regulatory networks
The level of mRNA expression in the cells was affected by miRNAs, through which the bindings of miRNAs with their dicers degrade mRNAs and weaken their translations. If the downregulated mRNAs in the NAc from mice with depression-like behaviors were caused by miRNAs, their corresponding miRNAs will be upregulated. To test this hypothesis and validate our data about mRNA changes, we performed a series of bioinformatics analyses.
The significantly changed miRNAs predicted target mRNAs well match the measures by mRNA sequencing based on the three databases (Targetscan, RNA22, and miRDB) about the complex interactions between miRNAs and mRNAs. Table 4 shows the alternated miRNAs and their predicted-target mRNAs. For instance, the unregulated mmu-miR-144-3p predicted the downregulated Gad1 and Nr3c1 as the target mRNAs. The altered mRNAs and their corresponding miRNAs are shown in Table 5. Among those predicted altered mRNAs, Bdnf and mRNAs could be predicted as the four changed miRNAs (mmu-let-7a-1-3p, mmu-miR-10a-5p, mmu-miR-206-3p, and mmu-miR-10b-5p). From Tables 1 and 3–5, we can find that the altered mRNAs and miRNAs were simultaneously and reversely changed. The consistent results by associatively sequencing mRNAs and miRNAs validated our analyses and strengthen our conclusion.
  | Table 5 The changed mRNAs are regulated by miRNAs |
miRNA-144-3p can bind to the GAD1 mRNA predicted region
To confirm the silico prediction, we selected miRNA-144-3p to examine whether they targeted Gad1 by qRT-PCR and dual luciferase reporter assay. Two areas of Gad1 3′-UTRs (769–775 and 902–908) were predicted as the miRNA-144-3p binding site according to the three databases (Figure 4A). The results of qRT-PCR analysis showed that there were negative correlations between miRNA-144-3p and Gad1 mRNA expression in NAc tissue from mice with depression-like behaviors and controls (r=-0.946; P<0.001; Figure 4B). Furthermore, in dual luciferase report assay, we constructed luciferase reporter plasmids, which contained the two wild-type or corresponding mutant 3′-UTRs of the predicted binding sites. These reporter constructs were transfected into HEK293T cells. After miRNA-144-3p mimics or its negative controls were incubated, the relative activities of luciferase reporter for the 3′-UTR 769–775 and 902–908 of Gad1 mRNA were significantly lowered by the mimics of miRNA-144-3p, which were reversed by mutating the binding sites of miRNA-144-3p. Interestingly, this phenomenon can also be eliminated by mutating both the binding sites of Gad1, which appeared to have the synergistic effect (Figure 4C). These results suggested that miR-144-3p might act as negative regulators of translation through binding to the two sites of Gad1 3′-UTR, which also supported our analyses for the prediction of miRNA target genes.
Discussion
MDD is a recurrent, devastating mental disorder, which affects >350 million people worldwide, and exerts substantial public health and financial costs to society. Thus, there is a significant need to discover innovative therapeutics to treat depression efficiently. Growing evidence indicates that dysfunctions in brain reward circuitry are major causes of mood disorders including depression. In the present study, we performed transcriptome analysis in CUMS model and reported differentially expressed genes and miRNAs in the NAc, a key brain region of reward circuitry.
The differentially expressed mRNAs were significantly enriched in GABAergic synapses, dopaminergic synapses, axon guidance, neurotrophin signaling pathway, neurotransmitter synthesis, autophagy-associated apoptotic pathway, and neural-immune process. It is noteworthy that the upregulated mRNAs were clustered in hemoglobin genes. In addition, target miRNAs and mRNAs were upregulated. The inline results from sequencing miRNAs and mRNAs (Tables 1 and 3) as well as qRT-PCR (Figures 2 and 3) fortify our results. Moreover, the consistent results of qRT-PCR and dual luciferase reporter assays also strengthen the bioinformatics analysis of miRNA target prediction (Figure 4). To the best of our knowledge, here for the first time, we reveal the molecular mechanism of the NAc pathophysiology in CUMS-induced depression.
Various research reports have highlighted the significance of NAc in MDD.12,31–35 In humans, deep brain stimulation of the NAc has been shown to alleviate anhedonic symptoms of depression.36 However, the NAc is a heterogeneous structure comprising different cell types, various dopamine receptors, and other neuromodulatory signaling. These characteristics made it difficult to elucidate the precise role of the NAc circuitry in depressive behaviors. Increasing evidence supports the reduction of synaptic transmission and GABAergic neurons in the NAc associated with depression.37–39 Vulnerable mice exhibit an elevated stubby spine NAc that is associated with high occurrence of miniature excitatory post-synaptic current.40 Electrophysiological record of NAc excitatory diffusion followed stress expose were unbalanced reduction.41 Also, in our previous study, we found that GABAergic neurons were attributed as inhibitory synapse outputs, excitatory, and excitability synapse reception in CUMS-induced depression mice.8 Such indications are completely in-line with our observations (Tables 1–5). As GABAergic neurons regulated the coordination and encoding ability of excitatory neurons,42,43 the attenuation of GABAergic neurons led to the dysfunction of neuronal networks in the NAc. Furthermore, our analyses indicated the functional defect in dopaminergic synapses, axon guidance, neurotrophin signaling pathway, and neuroactive ligand–receptor interaction. As was known to us, depression was characterized as low self-esteem and anhedonia, which might have occurred due to dysfunction of the synapses and neurons in the reward circuits, such as the synaptic transmission deficit and the degeneration of the axons in the NAc and medial PFC.44 Taken together, these results provided insight into mechanisms underlying the physiological absence of the reward circuits in psychiatric disorders for example, bipolar disorder, MDD, attention schizophrenia and post-traumatic stress disorder.
Earlier studies indicated downregulation in the genes of MAPK signaling pathway, neurotransmitter synthesis and myelin-associated protein in rodent animal model of depression-like and depressive patients with the use of qRT-PCR, microarray or in situ hybridization.45–49 These indications were also consistent with the current study in CUMS-induced depression-like mice. In addition, our analysis further implies autophagy-associated apoptotic pathway and neural-immune process, which may bring new insight into molecular mechanism essential depression. These latest findings might be due to the use of advanced mRNA sequencing method.
In this study, the leading observations were that the upregulated mRNAs were clustered in hemoglobin genes. Hemoglobin expression is high in erythrocytes. It has a key role in oxidative stress, response to injury, and neuronal respiration.50,51 Cells can use neuronal hemoglobin to produce hemoglobin derived peptides action on opioid and cannabin receptors.52 Chronic peripheral inflammation was contributed to increase the frontal expression of hemoglobin genes in ischemic and intracerebral hemorrhage patients.53,54 Upregulation of hemoglobin genes (Hbb-b1, Hba-a1, and Hba-a2) has also been found in the medial PFC of mice with CUMS-induced depression-like behaviors in our previous study.7 Human HbF and HbA2 were associated with disease progression in bipolar disorder, which had a shielding function of HbA2 against postpartum episodes.55 Thus, the elevation in hemoglobin genes was found in diverse pathological states. Consistent results of related studies reveal hemoglobin genes as the potential markers of chronic mild stress-induced depression.
miRNAs are a group of single-stranded small non-coding RNA molecules with 19–22 nucleotides, which could post-transcriptionally degrade mRNA or inhibit the translation of mRNA via binding the 3′-UTR section of mRNAs and furthermore influence the expression of target genes.56 In some cases, miRNAs might trigger translation or even control transcription level by binding to precise gene promoters. The role of miRNAs in depression is mainly explained from preclinical studies which showed the role of miRNAs in neurogenesis, neurodevelopment, and synaptic plasticity.57,58 Accumulated facts imply that stress is a significant risk factor in depression and can differentially regulate miRNA-mediated expression of mRNA transcripts in the brain of rodent.59–61 Additionally, mRNAs down-regulation in the mice NAc are rooted by miRNAs, and their related miRNAs will also be upregulated. To validate the hypothesis and strengthen our findings about mRNA changes, we investigated the miRNA expression in NAc by their sequencings from mice with depression-like behaviors.
In our current study, we found that some known miRNAs were significantly changed in mice with CUMS-induced depression-like behaviors (Table 1), which degraded mRNAs listed in Table 5. In other words, the analysis from miRNA sequencing was consistent with the analysis from mRNA sequencing. Furthermore, these dysregulation miRNAs were reported in previous studies by qRT-PCR, microarray, or situ hybridization in rodent depression animal model or depressive patients.62–68 We next performed a series of bioinformatics analyses for integrated miRNA/mRNA regulatory networks (Tables 4 and 5). For instance, the unregulated mmu-miR-144-3p predicted the downregulated Gad1 and Nr3c1 as the target mRNAs. Whereas, the downregulated mmu-miR-124-5p predicted the unregulated Aspa and Irf7 as the target mRNAs. Among those predicted altered mRNAs, Bdnf mRNA could be predicted as the four changed miRNAs (mmu-let-7a-1-3p, mmu-miR-10a-5p, mmu-miR-206-3p, and mmu-miR-10b-5p).
To verify the silico prediction, we selected miRNA-144-3p to test whether they targeted Gad1 by dual luciferase reporter assay. Our results demonstrated that miR-144-3p acted as negative regulators of translation through attaching with the two sites of Gad1 3′-UTR. miR-144-3p role still remained unclear in depressive disorders. But, some biological mechanisms can explain our judgments. miR-144-3p has extensive expression nature which is enriched in human brain, additionally, also enriched in malignant hematopoietic and normal cells and tissues.69 miR-144-3p is very conserved and has various envisaged targets in both mice and humans. Numerous investigations have revealed that miR-144-3p was implicated against response to stress, aging diseases, and mood stabilizer treatment.7,70,71 miR-144-3p targeting signaling pathways include Nrf2, Wnt/β-catenin, and MAKP pathways.72–74 In addition, miR-144-3p may suppress ataxin 1 (ATXN1) expression in human cells, and interestingly, the Genetic Association Database illustrated that ATXN1 is linked with psychological disorders.75 In terms of response to stress, miR-144-3p level is notably elevated in depressed patients compared with healthy young adults. These studies support that miR-144-3p-mediated target gene expressions were involved in the processes of the pathogenesis of depression.
The quantitative alterations of miRNAs in some reports without the alterations of their targeted mRNAs in others may be caused by the following reasons. The changes in miRNAs may not reach the threshold to regulate their targeted mRNAs.76 circRNAs act as miRNA sponges and positive regulators of miRNA-targeted genes.77 On the other hand, the quantitative changes of mRNAs without the alteration of their corresponding miRNAs imply that the altered mRNAs are likely regulated by other epigenetic mechanisms, such as DNA methylation and repressive histone modification in the promoters.78–80 Therefore, the associated analysis of miRNA/mRNAs in the brain regions with depression-related dysfunction may validate the data and strengthen the conclusion.
Conclusion
In summary, we performed a series of bioinformatics analyses for RNA sequencing data in the NAc tissue from the CUMS-induced depression mice. The worsening of dopaminergic synapses, GABAergic synapses, and neurotransmitter syntheses and autophagy-associated apoptotic pathway are associated with the molecular pathological mechanism of CUMS-induced depression. Our analyses support the string from stress to neuron atrophy through miRNA/mRNA regulatory network and provide the guidelines for developing novel therapeutic strategies for this complex disorder.
Acknowledgments
This work was supported by the Shandong Co-Innovation Center of Classic TCM Formula, Scientific Innovation Team of Shandong University of Traditional Chinese Medicine, the National Natural Science Foundation of China (81673852 and 81603419), Shandong Province University Scientific Research Project (J18KZ014), Shandong Medical Health Technology Development Plan Project (2018WS203), and Student Research Training Program of Shandong University of TCM (2018090 and 2018093).
Author contributions
All authors contributed toward data analysis, drafting and critically revising the paper, approved the final version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Anthes E. Depression: a change of mind. Nature. 2014;515(7526):185–187. | ||
Smith K. Mental health: a world of depression. Nature. 2014;515(7526):180–181. | ||
Gaynes BN, Warden D, Trivedi MH, Wisniewski SR, Fava M, Rush AJ. What did STAR*D teach us? Results from a large-scale, practical, clinical trial for patients with depression. Psychiatric Services. 2009;60(11):1439–1445. | ||
Ma K, Zhang H, Baloch Z. Pathogenetic and therapeutic applications of tumor necrosis factor-α (TNF-α) in major depressive disorder: a systematic review. Int J Mol Sci. 2016;17(5):733–751. | ||
Converge Consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523(7562):588–591. | ||
Price JL, Drevets WC. Neural circuits underlying the pathophysiology of mood disorders. Trends Cogn Sci. 2012;16(1):61–71. | ||
Ma K, Guo L, Xu A, Cui S, Wang J-H. Molecular mechanism for stress-induced depression assessed by sequencing miRNA and mRNA in medial prefrontal cortex. PLoS One. 2016;11(7):e0159093. | ||
Zhu Z, Wang G, Ma K, Cui S, Wang J-H. GABAergic neurons in nucleus accumbens are correlated to resilience and vulnerability to chronic stress for major depression. Oncotarget. 2017;8(22):35933–35945. | ||
Bopp SK, Heilbronner U, Schlattmann P, et al. Leptin gene polymorphisms are associated with weight gain during lithium augmentation in patients with major depression. Eur Neuropsychopharmacol. 2019;29(2):211–221. | ||
Kang H-J, Bae K-Y, Kim S-W, et al. Methylation of the glucocorticoid receptor gene associated with depression in patients with acute coronary syndrome. Psychoneuroendocrinology. 2019;101:42–49. | ||
Pan D, Xu Y, Zhang L, et al. Gene expression profile in peripheral blood mononuclear cells of postpartum depression patients. Sci Rep. 2018;8(1):10139. | ||
Knowland D, Lim BK. Circuit-based frameworks of depressive behaviors: the role of reward circuitry and beyond. Pharmacol Biochem Behav. 2018;174:42–52. | ||
Russo SJ, Nestler EJ. The brain reward circuitry in mood disorders. Nat Rev Neurosci. 2013;14(9):609–625. | ||
Francis TC, Lobo MK. Emerging role for nucleus accumbens medium spiny neuron subtypes in depression. Biol Psychiatry. 2017;81(8):645–653. | ||
Kravitz AV, Tye LD, Kreitzer AC. Distinct roles for direct and indirect pathway striatal neurons in reinforcement. Nat Neurosci. 2012;15(6):816–818. | ||
Anacker C, Scholz J, O’Donnell KJ, et al. Neuroanatomic differences associated with stress susceptibility and resilience. Biol Psychiatry. 2016;79(10):840–849. | ||
Chandra R, Lenz JD, Gancarz AM, et al. Optogenetic inhibition of D1R containing nucleus accumbens neurons alters cocaine-mediated regulation of Tiam1. Front Mol Neurosci. 2013;6:13. | ||
Chandra R, Francis TC, Konkalmatt P, et al. Opposing role for EGR3 in nucleus accumbens cell subtypes in cocaine action. J Neurosci. 2015;35(20):7927–7937. | ||
Francis TC, Chandra R, Friend DM, et al. Nucleus accumbens medium spiny neuron subtypes mediate depression-related outcomes to social defeat stress. Biol Psychiatry. 2015;77(3):212–222. | ||
Delaloye S, Holtzheimer PE. Deep brain stimulation in the treatment of depression. Dialogues Clin Neurosci. 2014;16(1):83–91. | ||
Friedman AK, Walsh JJ, Juarez B, et al. Enhancing depression mechanisms in midbrain dopamine neurons achieves homeostatic resilience. Science. 2014;344(6181):313–319. | ||
Fatemi SH, Folsom TD, Rooney RJ, Thuras PD. Expression of GABAA α2-, β1- and ε-receptors are altered significantly in the lateral cerebellum of subjects with schizophrenia, major depression and bipolar disorder. Transl Psychiatry. 2013;3(9):e303. | ||
Lopizzo N, Bocchio Chiavetto L, Cattane N, et al. Gene—“Environment Interaction in Major Depression: Focus on Experience-Dependent Biological Systems. Front Psychiatry. 2015;6(8):68. | ||
Maheu M, Lopez JP, Crapper L, Davoli MA, Turecki G, Mechawar N. MicroRNA regulation of central glial cell line-derived neurotrophic factor (GDNF) signalling in depression. Transl Psychiatry. 2015;5(2):e511. | ||
Saus E, Soria V, Escaramís G, et al. Genetic variants and abnormal processing of pre-miR-182, a circadian clock modulator, in major depression patients with late insomnia. Hum Mol Genet. 2010;19(20):4017–4025. | ||
Dwivedi Y, Roy B, Lugli G, Rizavi H, Zhang H, Smalheiser NR. Chronic corticosterone-mediated dysregulation of microRNA network in prefrontal cortex of rats: relevance to depression pathophysiology. Transl Psychiatry. 2015;5(11):e682. | ||
Xu A, Cui S, Wang J-H. Incoordination among subcellular compartments is associated with depression-like behavior induced by chronic mild stress. Int J Neuropsychopharmacol. 2016;19(5):pyv122. | ||
Tang J, Xue W, Xia B, et al. Involvement of normalized NMDA receptor and mTOR-related signaling in rapid antidepressant effects of Yueju and ketamine on chronically stressed mice. Scientific Reports. 2015;5(1):13573. | ||
Seo J-S, Wei J, Qin L, Kim Y, Yan Z, Greengard P. Cellular and molecular basis for stress-induced depression. Mol Psychiatry. 2017;22(10):1440–1447. | ||
Ma K, Xu A, Cui S, Sun M-R, Xue Y-C, Wang J-H. Impaired GABA synthesis, uptake and release are associated with depression-like behaviors induced by chronic mild stress. Transl Psychiatry. 2016;6(10):e910. | ||
Bagot RC, Parise EM, Peña CJ, et al. Ventral hippocampal afferents to the nucleus accumbens regulate susceptibility to depression. Nat Comm. 2015;6(1):7062. | ||
Nestler EJ. Role of the brain’s reward circuitry in depression: transcriptional mechanisms. Int Rev Neurobiol. 2015;124:151–170. | ||
Golden SA, Christoffel DJ, Heshmati M, et al. Epigenetic regulation of Rac1 induces synaptic remodeling in stress disorders and depression. Nat Med. 2013;19(3):337–344. | ||
Robison AJ, Vialou V, Sun H-S, et al. Fluoxetine epigenetically alters the CaMKIIα promoter in nucleus accumbens to regulate ΔFosB binding and antidepressant effects. Neuropsychopharmacology. 2014;39(5):1178–1186. | ||
Horschig JM, Smolders R, Bonnefond M, et al. Directed communication between nucleus accumbens and neocortex in humans is differentially supported by synchronization in the theta and alpha band. PLoS One. 2015;10(9):e0138685. | ||
Schlaepfer TE, Cohen MX, Frick C, et al. Deep brain stimulation to reward circuitry alleviates anhedonia in refractory major depression. Neuropsychopharmacology. 2008;33(2):368–377. | ||
Kohnomi S, Konishi S. Multiple actions of a D3 dopamine receptor agonist, PD128907, on GABAergic inhibitory transmission between medium spiny neurons in mouse nucleus accumbens shell. Neurosci Lett. 2015;600:17–21. | ||
Yu J, Yan Y, Li K-L, et al. Nucleus accumbens feedforward inhibition circuit promotes cocaine self-administration. Proc Nat Acad Sci. 2017;114(41):E8750–E8759. | ||
Adermark L, Söderpalm B, Burkhardt JM. Brain region specific modulation of ethanol-induced depression of GABAergic neurons in the brain reward system by the nicotine receptor antagonist mecamylamine. Alcohol. 2014;48(5):455–461. | ||
Christoffel DJ, Golden SA, Dumitriu D, et al. IκB kinase regulates social defeat stress-induced synaptic and behavioral plasticity. J Neurosci. 2011;31(1):314–321. | ||
Lim BK, Huang KW, Grueter BA, Rothwell PE, Malenka RC. Anhedonia requires MC4R-mediated synaptic adaptations in nucleus accumbens. Nature. 2012;487(7406):183–189. | ||
Luscher B, Fuchs T. GABAergic control of depression-related brain states. Advances in pharmacology. 2015;73:97–144. | ||
Gao S-F, Klomp A, Wu J-L, Swaab DF, Bao A-M. Reduced GAD(65/67) immunoreactivity in the hypothalamic paraventricular nucleus in depression: a postmortem study. J Affect Disord. 2013;149(1–3):422–425. | ||
Proulx CD, Hikosaka O, Malinow R. Reward processing by the lateral habenula in normal and depressive behaviors. Nat Neurosci. 2014;17(9):1146–1152. | ||
Elizalde N, Pastor PM, Garcia-García Álvaro L, et al. Regulation of markers of synaptic function in mouse models of depression: chronic mild stress and decreased expression of VGlut1. J Neurochem. 2010;19(5):1314. | ||
Malemud CJ, Miller AH. Pro-inflammatory cytokine-induced SAPK/MAPK and JAK/STAT in rheumatoid arthritis and the new anti-depression drugs. Expert Opin Ther Targets. 2008;12(2):171–183. | ||
Collins LM, Downer EJ, Toulouse A, Nolan YM. Mitogen-Activated Protein Kinase Phosphatase (MKP)-1 in nervous system development and disease. Mol Neurobiol. 2015;51(3):1158–1167. | ||
Lippiello P, Hoxha E, Speranza L, et al. The 5-HT7 receptor triggers cerebellar long-term synaptic depression via PKC-MAPK. Neuropharmacology. 2016;101:426–438. | ||
Kéri S, Szabó C, Kelemen O. Blood biomarkers of depression track clinical changes during cognitive-behavioral therapy. J Affect Disord. 2014;164:118–122. | ||
Richter F, Meurers BH, Zhu C, Medvedeva VP, Chesselet M-F. Neurons express hemoglobin alpha- and beta-chains in rat and human brains. J Comp Neurol. 2009;515(5):538–547. | ||
Schelshorn DW, Schneider A, Kuschinsky W, et al. Expression of hemoglobin in rodent neurons. J Cereb Blood Flow Metab. 2009;29(3):585–595. | ||
Gelman JS, Sironi J, Castro LM, Ferro ES, Fricker LD. Hemopressins and other hemoglobin-derived peptides in mouse brain: comparison between brain, blood, and heart peptidome and regulation in Cpefat/fat mice. J Neurochem. 2010;113(4):871–880. | ||
He Y, Hua Y, Liu W, Hu H, Keep RF, Xi G. Effects of cerebral ischemia on neuronal hemoglobin. J Cereb Blood Flow Metab. 2009;29(3):596–605. | ||
Sarlus H, Wang X, Cedazo-Minguez A, Schultzberg M, Oprica M. Chronic airway-induced allergy in mice modifies gene expression in the brain toward insulin resistance and inflammatory responses. J Neuroinflammation. 2013;10(1):99. | ||
Ince B, Guloksuz S, Altinbaş K, Oral ET, Alpkan LR, Altinoz MA. Minor hemoglobins HbA2 and HbF associate with disease severity in bipolar disorder with a likely protective role of HbA2 against post-partum episodes. J Affect Disord. 2013;151(1):405–408. | ||
Tavakolizadeh J, Roshanaei K, Salmaninejad A, et al. MicroRNAs and exosomes in depression: potential diagnostic biomarkers. J Cell Biochem. 2018;119(5):3783–3797. | ||
Dwivedi Y. microRNAs as biomarker in depression pathogenesis. Ann Psychiatry Ment Health. 2013;1(1):1003. | ||
Saavedra K, Molina-Márquez A, Saavedra N, Zambrano T, Salazar L. Epigenetic modifications of major depressive disorder. Int J Mol Sci. 2016;17(8):1279. | ||
Serafini G, Pompili M, Hansen KF, et al. The involvement of microRNAs in major depression, suicidal behavior, and related disorders: a focus on miR-185 and miR-491-3p. Cell Mol Neurobiol. 2014;34(1):17–30. | ||
Brites D, Fernandes A. Neuroinflammation and depression: microglia activation, extracellular microvesicles and microRNA dysregulation. Front Cell Neurosci. 2015;9:476. | ||
Maffioletti E, Tardito D, Gennarelli M, Bocchio-Chiavetto L. Micro spies from the brain to the periphery: new clues from studies on microRNAs in neuropsychiatric disorders. Front Cell Neurosci. 2014;8(82):75. | ||
Dwivedi Y. Emerging role of microRNAs in major depressive disorder: diagnosis and therapeutic implications. Dialogues in Clin Neurosci. 2014;16(1):43–61. | ||
Miao Z, Mao F, Liang J, Szyf M, Wang Y, Sun ZS. Anxiety-related behaviours associated with microRNA-206-3p and BDNF expression in pregnant female mice following psychological social stress. Mol Neurobiol. 2018;55(2):1097–1111. | ||
Yan H, Fang M, Liu X-Y. Role of microRNAs in stroke and poststroke depression. Sci World J. 2013;2013(4):1–6. | ||
Liu Q, Sun NN, Zz W, Fan DH, Cao MQ. Chaihu-Shugan-San exerts an antidepressive effect by downregulating miR-124 and releasing inhibition of the MAPK14 and GRIA3 signaling pathways. Neural Regener Res. 2018;13(5):837–845. | ||
Higuchi F, Uchida S, Yamagata H, et al. Hippocampal microRNA-124 enhances chronic stress resilience in mice. J Neurosci. 2016;36(27):7253–7267. | ||
Roy B, Dunbar M, Shelton RC, Dwivedi Y. Identification of MicroRNA-124-3p as a putative epigenetic signature of major depressive disorder. Neuropsychopharmacology. 2017;42(4):864–875. | ||
Hussain N, Zhu W, Jiang C, et al. Down-regulation of miR-10a-5p in synoviocytes contributes to TBX5-controlled joint inflammation. J Cell Mol Med. 2018;22(1):241–250. | ||
Pienimaeki-Roemer A, Konovalova T, Musri MM, et al. Transcriptomic profiling of platelet senescence and platelet extracellular vesicles. Transfusion. 2017;57(1):144–156. | ||
Liu B-B, Luo L, Liu X-L, Geng D, Liu Q, Yi L-T. 7-chlorokynurenic acid (7-CTKA) produces rapid antidepressant-like effects: through regulating hippocampal microRNA expressions involved in TrkB-ERK/Akt signaling pathways in mice exposed to chronic unpredictable mild stress. Psychopharmacology. 2015;232(3):541–550. | ||
Giridharan VV, Thandavarayan RA, Fries GR, et al. Newer insights into the role of miRNA a tiny genetic tool in psychiatric disorders: focus on post-traumatic stress disorder. Transl Psychiatry. 2016;6(11):e954. | ||
Lan F, Yu H, Hu M, Xia T, Yue X. miR-144-3p exerts anti-tumor effects in glioblastoma by targeting c-Met. J Neurochem. 2015;135(2):274–286. | ||
Feng Y, Li N, Ma H, Bei B, Han Y, Chen G. Undescribed phenylethyl flavones isolated from Patrinia villosa show cytoprotective properties via the modulation of the mir-144-3p/Nrf2 pathway. Phytochemistry. 2018;153:28–35. | ||
Li J, Sun P, Yue Z, Zhang D, You K, Wang J. miR-144-3p induces cell cycle arrest and apoptosis in pancreatic cancer cells by targeting proline-rich protein 11 expression via the mitogen-activated protein kinase signaling pathway. DNA and Cell Biology. 2017;36(8):619–626. | ||
Persengiev S, Kondova I, Otting N, Koeppen AH, Bontrop RE. Genome-wide analysis of miRNA expression reveals a potential role for miR-144 in brain aging and spinocerebellar ataxia pathogenesis. Neurobiol Aging. 2011;32(12):2316.e17-2. | ||
Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40(7):3131–3142. | ||
Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. | ||
Scala G, Marwah V, Kinaret P, Sund J, Fortino V, Greco D. Integration of genome-wide mRNA and miRNA expression, and DNA methylation data of three cell lines exposed to ten carbon nanomaterials. Data in Brief. 2018;19:1046–1057. | ||
Sijen T. Molecular approaches for forensic cell type identification: on mRNA, miRNA, DNA methylation and microbial markers. Forensic Sci Int Genet. 2015;18:21–32. | ||
Doecke JD, Wang Y, Baggerly K. Co-localized genomic regulation of miRNA and mRNA via DNA methylation affects survival in multiple tumor types. Cancer Genetics. 2016;209(10):463–473. |
Supplementary materials
  | Table S1 Chronic unpredictable mild stress procedure |
  | Table S2 qRT-PCR prime information |
  | Table S3 3′-Untranslated region (UTR) and site-directed mutation prime sequence Gad1 |
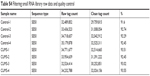  | Table S4 Filtering small RNA library raw data and quality control |
  | Table S5 Filtering transcriptome raw data and alignment |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.






