Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 13
Identification of Key Genes Involved in Diabetic Peripheral Neuropathy Progression and Associated with Pancreatic Cancer
Received 19 October 2019
Accepted for publication 1 February 2020
Published 19 February 2020 Volume 2020:13 Pages 463—476
DOI https://doi.org/10.2147/DMSO.S235011
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ming-Hui Zou
Liumeng Jian,1,* Guangda Yang2,*
1Department of Neurology, Zengcheng District People’s Hospital of Guangzhou, (BoJi-Affiliated Hospital of Sun Yat-Sen University), Guangzhou, People’s Republic of China; 2Department of Cancer Chemotherapy, Zengcheng District People’s Hospital of Guangzhou (BoJi-Affiliated Hospital of Sun Yat-Sen University), Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Guangda Yang
Department of Cancer Chemotherapy, Zengcheng District People’s Hospital of Guangzhou (BoJi-Affiliated Hospital of Sun Yat-Sen University), Guangzhou 511300, People’s Republic of China
Email [email protected]
Introduction: Diabetes mellitus (DM) patients suffer from high morbidity and premature mortality due to various diabetic complications and even cancers. Therefore, this study aimed to identify key genes involved in the pathogenesis of diabetic peripheral neuropathy (DPN) and pancreatic cancer (PC).
Methods: We analyzed three gene expression profiles (GSE95849, GSE28735 and GSE59953) to obtain differentially expressed genes (DEGs). Then, Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed by using the Database for Annotation, Visualization, and Integrated Discovery (DAVID). The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database was then used to establish a protein–protein interaction (PPI) network. The MCODE and cytoHubba plug-ins of Cytoscape were used to select hub genes. Finally, survival analysis of the hub genes was performed using the Kaplan–Meier plotter and GEPIA online tool.
Results: We first analyzed GSE95849 to obtain DPN-related genes. DEGs were obtained from three groups in GSE95849. The DEGs were enriched in the Toll-like receptor signaling pathway, hematopoietic cell lineage and chemokine signaling pathway. Importantly, we identified three shared genes as hub genes, including TLR4, CCR2 and MMP9. We then analyzed and integrated GSE95849 and GSE28735 to obtain genes common in DM and PC. A total of 58 mutual DEGs were identified, and these DEGs were enriched in the ECM-receptor interaction, focal adhesion and pathways in cancer. Five hub genes (including PLAU, MET, CLU, APOL1 and MMP9) were associated with the overall survival of PC patients. However, the results from the analysis of GSE59953 showed that hyperglycemia or TGF-β 1 treatment did not affect the expression level of these hub genes, but the DEGs based on hyperglycemia or TGF-β 1 treatment were mostly enriched in the ECM-receptor interaction, focal adhesion and pathways in cancer. Finally, functional enrichment analysis of MMP9 showed that significant genes correlated with MMP9 were associated with the tumorigenicity of cancers, insulin resistance, development of DM and inflammation.
Conclusion: In summary, inflammation and immunity-related pathways may play an important role in DM and DPN, while the ECM-receptor interaction, focal adhesion and pathways in cancer pathways may play significant roles in DM and PC. MMP9 may be used as a prognostic marker for PC and may be helpful for the treatment of DM, DPN and PC.
Keywords: diabetes mellitus, diabetic peripheral neuropathy, pancreatic cancer, GEO, MMP9
Introduction
Diabetes mellitus (DM) is one of the largest public health problems worldwide. Diabetic patients suffer from high morbidity and premature mortality due to the development of various diabetic complications, including cardiomyopathy, vasculopathy, nephropathy, retinopathy, and neuropathy.1 In addition, diabetes mellitus was found to be associated with increased pancreatic cancer risk.2
Diabetic peripheral neuropathy (DPN) is the most common diabetic complication, characterized by pain, paraesthesia and sensory loss, afflicting over 50% of people with diabetes.3 Previous studies have showed that the pathogenesis and underlying mechanisms leading to DPN are complex. Diabetes-related metabolic factors such as increased glucose, decreased insulin, and increased lipids produce changes underlying the development of diabetic neuropathy. Injury to neurons, microvascular endothelium, and Schwann cells in DM contributes to the pathogenesis of neuropathy. In addition, various stress pathways, including oxidative stress, inflammation, and apoptosis, are involved in the development of diabetic neuropathy.3–5 Crucial advances have been achieved in understanding the pathogenesis of DPN. However, controversy remains due to its multifactorial etiology.
Diabetes mellitus is associated with increased pancreatic cancer risk.2 A previous study revealed that the presence of diabetes mellitus significantly increased the risk of the subsequent development of pancreatic cancer by 2-fold.6 Interestingly, studies found that metformin use can prolong overall survival and lower mortality in patients with PC and DM.7,8 However, the underlying mechanisms and potential interconnections between diabetes and pancreatic cancer are not fully well known.
Therefore, we performed this bioinformatics analysis by using data from the GEO database. We first identified the key genes and pathways involved in DPN and then analyzed the key genes and pathways involved in both DM and PC. Interestingly, we identified MMP9 as the common hub gene of DM, DPN and PC. The analysis results will provide meaningful clues for the treatment of DM, DPN and PC.
Materials and Methods
Microarray Data Information and DEGs Identification
GSE95849, GSE28735 and GSE59953 gene expression profiles were downloaded from the GEO database. DEGs were obtained using the web tool GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/). The data set GSE958499 consisted of 6 Diabetic peripheral neuropathy (DPN), 6 diabetes mellitus (DM), and 6 healthy controls (CN). To obtained DEGs associated with DPN, the conditions of |log FC| ≥ 2, P-value< 0.05 was used for DM vs CN and DPN vs CN group. While |log FC| ≥1.5, P-value< 0.05 was used for DPN vs DM group. GSE2873510,11 contained 45 pairs of pancreatic cancer (PC) and adjacent non-tumor tissues. To obtained DEGs associated with DM and PC, the cutoff criterion of |log FC| ≥1, P-value< 0.05 was used for DM vs CN group of GSE95849 and PC vs CN group of GSE28735. Then, venn diagram was composed to select mutual DEGs between GSE95849 and GSE28735. GSE5995312 contained 4 samples of pancreatic stellate cells (PSCs) that cultured in normal glucose concentration (control), 4 samples of PSCs that exposed to 21 d hyperglycemia (21 d treatment), 4 samples of PSCs that treated with 48 h TGF-β1 treatment (48 h treatment) and 4 samples of PSCs that exposed to 21 d hyperglycemia + subsequent TGF-β1 treatment (21 d + 48 h treatment). To obtained DEGs, the conditions of |log FC| ≥ 0.5, P-value< 0.05 was used for control vs 21 d treatment group and control vs 48 h treatment group. While |log FC| ≥1.5, P-value< 0.05 was used for control vs 21 d + 48 h treatment group.
Genetic Ontology and Pathway Enrichment Analysis of DEGs
In order to explore the biological functional roles of the above DEGs, the DAVID was used to GO and KEGG pathway enrichment analysis. P<0.05 was used as the cutoff criterion.
Establishment of PPI Network and Modules Selection
The STRING database was used to construct PPI network. The Cytoscape software (http://www.cytoscape.org/) was used to visualize and analyze the PPI network. The Molecular Complex Detection (MCODE) plug-in Cytoscape was used to select the significant modules.
Identification of Hub Genes
The degrees method of cytoHubba or the MCODE scores of the Cytoscape software was used to select important hub genes among DEGs. For DEGs about DPN, the top 30 genes with high degrees or MCODE scores ≥12 were taken as the cutoff criterion. A Venn diagram was composed to select mutual genes from two methods. The mutual genes from the three groups were selected as the hub genes. For DEGs between DM and PC, the top 15 genes with high degrees were taken as the hub genes. For DEGs from GSE59953, the top 30 genes with high degrees were showed.
Survival Analysis
The Kaplan–Meier plotter (http://kmplot.com/analysis/index.php?p=background)13 and the Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer‐pku.cn/)14 databases were used for survival analysis of the hub genes. GEPIA was also used for expression levels analysis of the hub genes.
LinkedOmics Analysis
The LinkedOmics database (http://www.linkedomics.org/login.php)15 contains multi-omics data and clinical data for 32 cancer types and a total of 11,158 patients from The Cancer Genome Atlas (TCGA) project. The LinkFinder module of LinkedOmics was used to analyze genes differentially expressed in correlation with MMP9 in the TCGA PC cohort (n=178). Results were analyzed using Spearman correlation coefficient. The Link-Interpreter module of LinkedOmics was used to perform pathway and network analysis of differentially expressed genes. Gene set enrichment analysis (GSEA) was used to perform GO and KEGG pathway analysis of the data from the LinkFinder results. The rank criterion was a false discovery rate (FDR) < 0.05.
Results
Identification of DEGs Among Different Groups of GEO Data Sets
In the present study, identification of key genes and pathways involved in DPN, DM and PC were performed by integrated bioinformatics analysis (Figure 1). We first identified DEGs among different groups of GSE95849, which contained 6 DPN, 6 DM, and 6 CN samples. For DM vs CN group, 497 DEGs were screened. While a total of 1362 DEGs were screened from DPN vs CN group and 842 DEGs from DPN vs DM group (Figure 1). We second analyzed and integrated GSE95849 and GSE28735 to identify genes between DM and PC. A total of 3739 DEGs were selected from GSE95849 and 413 DEGs from GSE28735 (Figure 1).
GO and KEGG Pathway Analysis of DEGs Among Different Groups in GSE95849
We then performed GO and KEGG pathway analysis of the above DEGs by using DAVID. For DM vs CN group, the DEGs were enriched in defense response, inflammatory response, response to wounding, immune response and chemotaxis in the biological process (BP) category. Moreover, these genes were significantly enriched in the Golgi apparatus, cell fraction, cytoplasmic vesicle, vesicle and membrane fraction in the cellular component (CC) category. They were enriched in cytokine binding, protein domain-specific binding, chemokine receptor activity, chemokine binding and GTPase binding in the molecular function (MF) category (Table 1, left column). For DPN vs CN group, the DEGs were enriched in inflammatory response, response to wounding, defense response, immune response and innate immune response in the BP category. For the CC category, the DEGs were enriched in the Golgi apparatus, cell fraction, cytoplasmic vesicle, vesicle and membrane fraction. For the MF category, the DEGs were enriched in cytokine binding, protein domain-specific binding, chemokine receptor activity, chemokine binding and GTPase binding (Table 1, middle column). For DPN vs DM group, the DEGs were enriched in response to wounding, behavior, inflammatory response, cell activation and defense response in the BP category. With regard to the CC category, the DEGs were enriched in cell fraction, axon part, cell surface, vesicle membrane and soluble fraction. For the MF category, the DEGs were enriched in protein complex binding, chemokine receptor binding, chemokine activity, transcription factor binding and transcription factor activity (Table 1, right column).
 |
Table 1 Functional Enrichment Analysis for the DEGs of Different Groups of GSE95849 |
KEGG pathway enrichment analysis showed that the DEGs were enriched in the cytokine-cytokine receptor interaction, Toll-like receptor signaling pathway, hematopoietic cell lineage, PPAR signaling pathway and chemokine signaling pathway in the DM vs CN group (Table 2, up column). With regard to the DPN vs CN group, the DEGs were enriched in the Toll-like receptor signaling pathway, apoptosis, hematopoietic cell lineage, citrate cycle (TCA cycle), the NOD-like receptor signaling pathway, the chemokine signaling pathway, the MAPK signaling pathway and the B cell receptor signaling pathway (Table 2, middle column). For the DPN vs DM group, the DEGs were enriched in the NOD-like receptor signaling pathway, the MAPK signaling pathway, the neurotrophin signaling pathway, the B cell receptor signaling pathway, the chemokine signaling pathway, hematopoietic cell lineage, the Toll-like receptor signaling pathway, citrate cycle (TCA cycle) and cytokine-cytokine receptor interaction (Table 2, bottom column). The Toll-like receptor signaling pathway, hematopoietic cell lineage and chemokine signaling pathway were shown in all three groups. Therefore, these three signaling pathways may play an important role in DM and DPN.
 |
Table 2 The KEGG Pathway Enrichment Analysis for the DEGs of Different Groups of GSE95849 |
PPI Network Analysis and Screening for Hub Genes
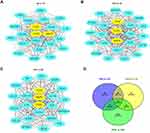
We further imported the above DEGs into STRING to obtain a PPI network. The interaction network was then imported into Cytoscape to screen the hub genes using two methods, including the degree method scores (Degrees) and MCODE. First, the cytoHubba plug-in of the Cytoscape software was used to select genes with the highest degrees. The top 30 genes with the highest degrees of connectivity for each group are shown in Table 3. Second, the MCODE plug-in of Cytoscape was used to select the most significant modules (Figure 2A–C). The genes with MCODE score ≥ 12 in each of the three modules are shown in Table 3. The mutual genes from the two methods were selected for each group. In the end, the Venn diagram identified three overlapping genes as representative hub genes from the three groups (Figure 2D), including TLR4, CCR2 and MMP9. Therefore, these hub genes may serve as promising biomarkers of DM and DPN.
 |
Table 3 Identified of the Hub Genes of Different Groups of GSE95849 by Degree and MCODE-Score of Cytoscape Sofeware |
Identification of Hub Genes Between DM and PC
DM has been identified as a risk factor for PC. We therefore tried to identify hub genes between DM and PC by analyzing the data set GSE95849 and GSE28735 datasets. A total of 3739 DEGs were screened between DM and CN in GSE95849, and 413 DEGs were screened between PC and CN in GSE28735. The 59 mutual DEGs were found via a Venn diagram (Figure 3A) and were imported into DAVID to perform GO and KEGG pathway analysis (Table S1). Functional analysis showed that the mutual DEGs were enriched in response to wounding, cell adhesion, biological adhesion, cell migration, immune response, defense response, inflammatory response, wound healing and blood vessel morphogenesis in the BP category. For the CC category, the mutual DEGs were enriched in the extracellular region, membrane fraction, insoluble fraction, cell fraction, extracellular region part, proteinaceous extracellular matrix, extracellular matrix, extracellular space, extracellular matrix part, platelet alpha granule, integral to plasma membrane, intrinsic to plasma membrane and basement membrane. With regard to the MF category, the mutual DEGs were enriched in glycosaminoglycan binding, hyaluronic acid binding, polysaccharide binding, pattern binding, growth factor binding, carbohydrate binding, vitamin binding and amine transmembrane transporter activity. KEGG pathway analysis showed that the mutual DEGs were enriched in ECM-receptor interaction, focal adhesion, pathways in cancer and the adipocytokine signaling pathway (Table S1). These results indicated that the three pathways may play an important role in DM and PC.
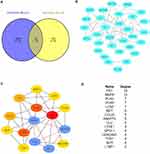
The 59 mutual DEGs were then imported into STRING to construct a PPI network, which had 31 nodes and 64 edges (Figure 3B). The interaction network was then imported into Cytoscape to screen the hub genes using the Degree algorithm. The interaction network with the top 15 genes with the highest degrees was created and is shown in Figure 3C. The degrees of the top 15 genes (including FN1, MMP9, PLAU, VCAN, LCN2, MET, CCL20, ANGPT2, CLU, LYVE1, APOL1, CEACAM1, TCN1, SLP1 and LTBP1) are shown in Figure 3D. Interestingly, the significant module generated by MCODE contained four genes, including MMP9, PLAU, MET and ANGTP2 (Figure 3C).
We then used the GEPIA and Kaplan–Meier plotter online databases to analyze the expression level and prognostic value of the 15 hub genes. The analysis showed that only five hub genes (including PLAU, MET, CLU, APOL1 and MMP9) were associated with the overall survival of PC patients (Figure S1B, C, E, F, H, I, K, L and Figure 4B and C). The expression of PLAU, MET, APOL1 and MMP9 was significantly higher in PC tissues than in paired normal tissues (Figure S1A, D, G, J and Figure 4A). However, the expression level of CLU was not significantly different between PC tissues and paired normal tissues (Figure S1G). These results indicated that the five hub genes may serve as promising biomarkers of DM and PC.
Diabetes mellitus was associated with increased pancreatic cancer risk. In addition, a study found that elevated fasting plasma glucose (FPG) levels are risk factors for PC.16 A study found that activated human pancreatic stellate cells (PSCs) are the major cellular constituents of the pancreatic ductal adenocarcinoma stroma.17 Katalin Kiss et al kept PSCs under chronic hyperglycemic conditions; the PSCs were subsequently either treated or untreated with TGFβ1 to model the gene expression changes, and the GSE59953 dataset was generated with this data.12 In vivo, they found that chronic hyperglycemia induced the transdifferentiation of PSCs and enhanced the malignant molecular communication with PSCs.12 Therefore, we used GSE59953 dataset to analyze the effect of chronic hyperglycemia (CHG) on gene expression. For the control vs 21 d treatment group, a total of 854 DEGs were identified. For the control vs 48 h treatment group, a total of 205 DEGs were identified. For the control vs 21 d + 48 h treatment group, a total of 1167 DEGs were identified. KEGG pathway analysis showed that the DEGs were also mostly enriched in the ECM-receptor interaction, focal adhesion and pathways in cancer (Table S2). We also constructed a PPI network and selected the significant module to identify hub genes (Figure S2A–C and Table S3). In the end, the Venn diagram identified six overlapping genes (CXCL8, EGR1, FN1, FOS, PPARG and SPP1) as representative hub genes from the three groups (Figure S2D). Unfortunately, the hub genes identified from GSE95849 and GSE28735 were not significantly altered under hyperglycemic conditions. However, they may share similar pathways, such as the ECM-receptor interaction, focal adhesion and pathways in cancer.
Enrichment Analysis of MMP9 Functional Networks in PC
The comprehensive results revealed that MMP9 was found to be both involved in DPN progression and was associated with PC. We used the function module of LinkedOmics to analyze MMP9 mRNA sequencing data from 178 PC patients in the TCGA. As shown in the volcano plot in Figure 5A, MMP9 was positively correlated with 1550 genes (dark red dots) but negatively correlated with 729 genes (dark green dots) (FDR < 0.01). The 50 significant gene sets positively and negatively associated with MMP9 are shown in the heat map in Figure 5B and C. The top 20 significant genes positively and negatively associated with MMP9 are shown more detail in Table S4. MMP9 expression showed a strong positive or negative association with the expression of C1QTNF6 (Spearman correlation = 0.547, p-value= 1.00e–15), TWIST1 (Spearman correlation = 0.542, p-value = 1.00e–15), TGFB1(Spearman correlation =0.523, p-value =1.00E-15), IL4I1 (Spearman correlation =0.513, p-value =1.00E-15), OSCAR (Spearman correlation =0.498, p-value =1.00E-15), CASC2 (Spearman correlation = −0.449, p-value =4.98E-10), PTPRN2 (Spearman correlation =−0.422, p-value =6.12E-09), KIF3B (Spearman correlation =−0.417, p-value =7.05E-09) and DCLRE1A (Spearman correlation =−0.407, p-value =1.67E-08), which were associated with the tumorigenicity of cancers, insulin resistance, development of diabetes mellitus and inflammation. KEGG pathway analysis by GSEA showed that the genes correlated with MMP9 were significantly enriched in cytokine-cytokine receptor interaction (FDR=0, p-value=0), ECM-receptor interaction (FDR=0, p-value=0), hematopoietic cell lineage (FDR=0, p-value=0), B cell receptor signaling pathway (FDR=0, p-value=0), citrate cycle (TCA cycle) (FDR=0, p-value=0) and focal adhesion (FDR=0, p-value=0) (Figure 5D–J). Therefore, MMP9 has the potential to be used as a target for PC diagnosis and treatment.
Discussion
In the present study, we performed bioinformatics analysis for two purposes. One was to identify the key genes and biological pathways responsible for the progression of DPN. The other purpose was to identify the key genes and biological pathways between DM and PC. By using the GSE95849 dataset, functional enrichment analysis showed that most of the DEGs from the three compared groups (DM vs CN, DPN vs CN and DPN vs DM) were enriched in defense response, inflammatory response, immune response and response to wounding. KEGG pathway analysis found that the DEGs from the three compared groups shared three pathways, including the Toll-like receptor signaling pathway, hematopoietic cell lineage and chemokine signaling pathway. Moreover, in comparison with DM, the DPN-specific DEGs were also significantly enriched in the NOD-like receptor signaling pathway, MAPK signaling pathway, neurotrophin signaling pathway, B cell receptor signaling pathway, citrate cycle (TCA cycle) and cytokine-cytokine receptor interaction. Interestingly, most of these pathways were associated with inflammation and immune-related functions. Therefore, inflammation and immune-related pathways may play an important role in DM and DPN.
Importantly, we identified three hub genes, including TLR4, CCR2 and MMP9. TLR4 belongs to the Toll-like receptor (TLR) family, which plays an important role in the innate immune response.18 In addition, TLR4 was found to play a critical role in the pathophysiological process of diabetes19 and was identified as a potential diagnostic biomarker for DPN.20 CC chemokine receptor 2 (CCR2) plays an important role in the recruitment and activation of macrophages and in turn contributes to both inflammatory and neuropathic pain states.21 CCR2 plays a critical role in a range of inflammatory diseases in the central nervous system (CNS).21 In addition, chemokine C-C motif ligand 2 (CCL2)-CCR2 signaling was involved in the maintenance of orofacial neuropathic pain.22 Blockade of the CCL2/CCR2 pathway improves diabetic nephropathy.23 Pharmacological modulation of the CCL2/CCR2 pathway results in the attenuation of neuropathic pain.24 Evidence has shown that matrix metalloproteinases (MMPs) can cleave multiple substrates of the CNS and contribute to promoting and dampening diseases of the CNS.25,26 Moreover, MMPs are involved in diabetes-induced retinal neuropathy.27 In addition, MMPs can degrade extracellular matrix (ECM) proteins, which in turn results in tissue remodeling and can contribute to cell migration.28,29 Moreover, MMP9 was found to be involved in the pathogenesis of DM and diabetic complications such as diabetic retinopathy.30 Therefore, these hub genes may serve as promising biomarkers of DM and DPN.
Another important finding is that we identified 15 hub genes between DM and PC by analyzing the GSE95849 and GSE28735 datasets. Five genes (MMP9, PLAU, MET, CLU and APOL1) were associated with the overall survival of PC. These results indicated that they may be identified as biomarkers and therapeutic targets of PC by further studies. We then focused on MMP9, as it was also identified as the key gene involved in DPN in the present study. We used the LinkedOmics database to conduct the functional enrichment analysis of MMP9 in PC. KEGG pathway analysis showed that the genes correlated with MMP9 were significantly enriched in cytokine-cytokine receptor interaction, ECM-receptor interaction, hematopoietic cell lineage, B cell receptor signaling pathway, citrate cycle (TCA cycle) and focal adhesion. Interestingly, KEGG pathway analysis of the 59 mutual DEGs showed that they were also significantly enriched in the ECM-receptor interaction and focal adhesion. In addition, MMPs can degrade extracellular matrix (ECM) proteins, which can in turn result in tissue remodeling and contribute to cell migration.28,29 MMPs were found to be involved in immunity and inflammation by regulating cytokine and chemokine function.31 Anti–inflammatory macrophages enhance the invasion of pancreatic cancer by inducing MMP9 and ADAM8 expression.32 In addition, MMP-9 expression was associated with poorer prognosis of pancreatic cancer.33 A previous study has found that the inhibition of MMPs in vitro can reduce metastases of pancreatic cancer.34 Interestingly, MMP-9 has been identified as an activator of nanosystems for targeted drug delivery in pancreatic cancer.35 In the preclinical models of pancreatic cancer, the addition of an anti-MMP9 antibody can prolong animal survival.36 These results may suggest that the MMP9 antibody could be exploited to improve clinical PC therapy. Moreover, most of the significant genes that were correlated with MMP9, such as C1QTNF6, TWIST, HK3, CASC2, TGF-β1 and OSCAR, were associated with the tumorigenicity of cancers, insulin resistance, development of diabetes mellitus and inflammation. C1q/tumor necrosis factor-related protein-6 (C1QTNF6, also known as CTRP6) has been found to be associated with insulin resistance and the development of diabetes in the Chinese population.37 CTRP6 has been identified as an important metabolic/immune regulator linking obesity to adipose tissue inflammation and insulin resistance.38 A study found that C1QTNF6 was upregulated in gastric carcinoma and contributed to the migration and proliferation of gastric carcinoma cells.39 Study showed that targeting twist family bHLH transcription factor 1 (TWIST1) through loss of function can inhibit the tumorigenicity of glioblastoma.40 High hexokinase 3 (HK3) expression was associated with epithelial-mesenchymal transition (EMT) in colorectal cancer.41 High levels of TGF-β1 were associated with the susceptibility to type 2 diabetes mellitus and type 2 diabetic nephropathy.42 In addition, stimulation with TGF-beta signaling can enhance stem cell properties in colorectal cancer.43 Upregulation of the lncRNA cancer susceptibility 2 (CASC2) suppressed the cell proliferation and metastasis of breast cancer via the inactivation of the TGF-β signaling pathway.44 lncRNA CASC2 expression was downregulated in patients with type 2 diabetes combined with chronic renal failure compared with healthy controls.45 The osteoclast associated Ig-like receptor (OSCAR)-surfactant protein D interaction can activate TNF-α release from human CCR2+ inflammatory monocytes46 and OSCAR-collagen signaling in monocytes plays a pro-inflammatory role and contributes to the pathogenesis of rheumatoid arthritis.47 These data indicated that MMP9 has the potential to be used as a target for the diagnosis and treatment of DM and PC.
Although bioinformatics technologies have the potential to identify candidate genes in various diseases, some limitations remain in this study. First, the RNA samples in GSE95849 were obtained from peripheral blood mononuclear cells (PBMCs) and not from peripheral nerves, so it may be difficult to make the conclusion that these hub genes contributed to the development of DPN. However, it is very difficult to conclude the collection of nerve biopsies in studies of patients with diabetic neuropathy. Transcriptome studies in PBMCs have emgeted as a new kind of transcriptional analysis and have been widely used in diabetic studies.48,49 Therefore, the results analyzed from this dataset are dependable. Second, the samples in GSE95849 and GSE28735 were obtained from DM and PC, respectively. Therefore, the linked between DM and PC was not directly assessed. Third, the samples in GSE59953 were from PSCs; thus, it is also difficult to fully reflect the real word. Furthermore, further biological experiments are needed to validate our results because our study was performed by bioinformatics analysis based on GEO datasets.
However, our data provide a comprehensive bioinformatics analysis of the DEGs and pathways that may be involved in DPN, DM and PC. We further confirmed that inflammation, immunity and their signaling pathways play an important role in DM and DPN. The ECM-receptor interaction, focal adhesion and pathways in cancer may play significant roles in DM and PC. More importantly, we identified MMP9 as the most important hub gene, which served as an inflammatory/immune regulator in DPN and linked DM to pancreatic cancer. We propose that careful targeting of MMP9 could be helpful for the treatment of DM, DPN and PC, but more studies are needed.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
We thank all the doctors in our department for their help in patients care so that we can have enough time to conduct this study.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Forbes JM, Cooper ME. Mechanisms of diabetic complications. Physiol Rev. 2013;93(1):137–188. doi:10.1152/physrev.00045.2011
2. Andersen DK, Andren-Sandberg Å, Duell EJ, et al. Pancreatitis-diabetes-pancreatic cancer. Pancreas. 2013;42(8):1227–1237. doi:10.1097/MPA.0b013e3182a9ad9d
3. Edwards JL, Vincent AM, Cheng HT, Feldman EL. Diabetic neuropathy: mechanisms to management. Pharmacol Therapeut. 2008;120(1):1–34. doi:10.1016/j.pharmthera.2008.05.005
4. Vincent AM, Calabek B, Roberts L, Feldman EL. Biology of diabetic neuropathy. Handb Clin Neurol. 2013;115:591.
5. Vincent AM, Callaghan BC, Smith AL, Feldman EL. Diabetic neuropathy: cellular mechanisms as therapeutic targets. Nat Rev Neurol. 2011;7(10):573–583. doi:10.1038/nrneurol.2011.137
6. Stolzenberg-Solomon RZ, Graubard BI, Chari S, et al. Insulin, glucose, insulin resistance, and pancreatic cancer in male smokers. JAMA. 2005;294(22):2872–2878. doi:10.1001/jama.294.22.2872
7. Choi Y, Kim T, Oh D, et al. The impact of diabetes mellitus and metformin treatment on survival of patients with advanced pancreatic cancer undergoing chemotherapy. Cancer Res Treat. 2016;48(1):171–179. doi:10.4143/crt.2014.292
8. Sadeghi N, Abbruzzese JL, Yeung SCJ, Hassan M, Li D. Metformin use is associated with better survival of diabetic patients with pancreatic cancer. Clin Cancer Res. 2012;18(10):2905–2912. doi:10.1158/1078-0432.CCR-11-2994
9. Luo L, Zhou WH, Cai JJ, et al. Gene expression profiling identifies downregulation of the neurotrophin-mapk signaling pathway in female diabetic peripheral neuropathy patients. J Diabetes Res. 2017;2017:8103904. doi:10.1155/2017/8103904
10. Zhang G, Schetter A, He P, et al. DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity and predicts clinical outcome in pancreatic ductal adenocarcinoma. PLoS One. 2012;7(2):e31507. doi:10.1371/journal.pone.0031507
11. Zhang G, He P, Tan H, et al. Integration of metabolomics and transcriptomics revealed a fatty acid network exerting growth inhibitory effects in human pancreatic cancer. Clin Cancer Res. 2013;19(18):4983–4993. doi:10.1158/1078-0432.CCR-13-0209
12. Kiss K, Baghy K, Spisák S, et al. Chronic hyperglycemia induces trans-differentiation of human pancreatic stellate cells and enhances the malignant molecular communication with human pancreatic cancer cells. PLoS One. 2015;10(5):e128059. doi:10.1371/journal.pone.0128059
13. Nagy Á, Lánczky A, Menyhárt O, Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep-Uk. 2018;8(1):9227.
14. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
15. Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–D963. doi:10.1093/nar/gkx1090
16. Liao WC, Tu YK, Wu MS, Lin JT, Wang HP, Chien KL. Blood glucose concentration and risk of pancreatic cancer: systematic review and dose-response meta-analysis. BMJ. 2015;350:g7371. doi:10.1136/bmj.g7371
17. Fujita H, Ohuchida K, Mizumoto K, et al. alpha-smooth muscle actin expressing stroma promotes an aggressive tumor biology in pancreatic ductal adenocarcinoma. Pancreas. 2010;39(8):1254–1262. doi:10.1097/MPA.0b013e3181dbf647
18. Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4(7):499–511. doi:10.1038/nri1391
19. Dasu MR, Devaraj S, Park S, Jialal I. Increased toll-like receptor (TLR) activation and TLR ligands in recently diagnosed Type 2 diabetic subjects. Diabetes Care. 2010;33(4):861–868. doi:10.2337/dc09-1799
20. Zhu T, Meng Q, Ji J, Lou X, Zhang L. Toll-like receptor 4 and tumor necrosis factor-alpha as diagnostic biomarkers for diabetic peripheral neuropathy. Neurosci Lett. 2015;585:28–32. doi:10.1016/j.neulet.2014.11.020
21. Chu HX, Arumugam TV, Gelderblom M, Magnus T, Drummond GR, Sobey CG. Role of CCR2 in inflammatory conditions of the central nervous system. J Cereb Blood Flow Metab. 2014;34(9):1425–1429. doi:10.1038/jcbfm.2014.120
22. Zhang Z, Dong Y, Lu Y, Cao S, Zhao Z, Gao Y. Chemokine CCL2 and its receptor CCR2 in the medullary dorsal horn are involved in trigeminal neuropathic pain. J Neuroinflamm. 2012;9(1):136. doi:10.1186/1742-2094-9-136
23. Seok SJ, Lee ES, Kim GT, et al. Blockade of CCL2/CCR2 signalling ameliorates diabetic nephropathy in db/db mice. Nephrol Dial Transpl. 2013;28(7):1700–1710. doi:10.1093/ndt/gfs555
24. Piotrowska A, Kwiatkowski K, Rojewska E, et al. Direct and indirect pharmacological modulation of CCL2/CCR2 pathway results in attenuation of neuropathic pain — in vivo and in vitro evidence. J Neuroimmunol. 2016;297:9–19. doi:10.1016/j.jneuroim.2016.04.017
25. Rivera S. Metalloproteinases in nervous system function and pathology: introduction. Cell Mol Life Sci. 2019;76(16):3051–3053. doi:10.1007/s00018-019-03172-8
26. Chopra S, Overall CM, Dufour A. Matrix metalloproteinases in the CNS: interferons get nervous. Cell Mol Life Sci. 2019;76(16):3083–3095. doi:10.1007/s00018-019-03171-9
27. Opdenakker G, Abu El-Asrar A. Metalloproteinases mediate diabetes-induced retinal neuropathy and vasculopathy. Cell Mol Life Sci. 2019;76(16):3157–3166. doi:10.1007/s00018-019-03177-3
28. Wolf K, Te Lindert M, Krause M, et al. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol. 2013;201(7):1069–1084. doi:10.1083/jcb.201210152
29. Bourboulia D, Stetler-Stevenson WG. Matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases (TIMPs): positive and negative regulators in tumor cell adhesion. Semin Cancer Biol. 2010;20(3):161–168. doi:10.1016/j.semcancer.2010.05.002
30. Mishra M, Flaga J, Kowluru RA. Molecular mechanism of transcriptional regulation of matrix metalloproteinase-9 in diabetic retinopathy. J Cell Physiol. 2016;231(8):1709–1718. doi:10.1002/jcp.v231.8
31. Chakrabarti S, Patel KD. Matrix metalloproteinase-2 (MMP-2) and MMP-9 in pulmonary pathology. Exp Lung Res. 2009;31(6):599–621. doi:10.1080/019021490944232
32. Puolakkainen P, Koski A, Vainionpää S, et al. Anti-inflammatory macrophages activate invasion in pancreatic adenocarcinoma by increasing the MMP9 and ADAM8 expression. Med Oncol. 2014;31(3). doi:10.1007/s12032-014-0884-9
33. Xu Y, Li Z, Jiang P, et al. The co-expression of MMP-9 and Tenascin-C is significantly associated with the progression and prognosis of pancreatic cancer. Diagn Pathol. 2015;10(1). doi:10.1186/s13000-015-0445-3
34. Jimenez RE, Hartwig W, Antoniu BA, Compton CC, Warshaw AL, Fernandez-Del CC. Effect of matrix metalloproteinase inhibition on pancreatic cancer invasion and metastasis: an additive strategy for cancer control. Ann Surg. 2000;231(5):644–654. doi:10.1097/00000658-200005000-00004
35. Grunwald B, Vandooren J, Locatelli E, et al. Matrix metalloproteinase-9 (MMP-9) as an activator of nanosystems for targeted drug delivery in pancreatic cancer. J Control Release. 2016;239:39–48. doi:10.1016/j.jconrel.2016.08.016
36. Awasthi N, Mikels Vigdal AJ, Stefanutti E, et al. Therapeutic efficacy of anti-MMP9 antibody in combination with nab-paclitaxel-based chemotherapy in pre‐clinical models of pancreatic cancer. J Cell Mol Med. 2019;23(6):3878–3887. doi:10.1111/jcmm.2019.23.issue-6
37. Wang M, Tang X, Li L, et al. C1q/TNF-related protein-6 is associated with insulin resistance and the development of diabetes in Chinese population. Acta Diabetol. 2018;55(12):1221–1229. doi:10.1007/s00592-018-1203-2
38. Lei X, Seldin MM, Little HC, Choy N, Klonisch T, Wong GW. C1q/TNF-related protein 6 (CTRP6) links obesity to adipose tissue inflammation and insulin resistance. J Biol Chem. 2017;292(36):14836–14850. doi:10.1074/jbc.M116.766808
39. Qu HX, Cui L, Meng XY, et al. C1QTNF6 is overexpressed in gastric carcinoma and contributes to the proliferation and migration of gastric carcinoma cells. Int J Mol Med. 2019;43(1):621–629. doi:10.3892/ijmm.2018.3978
40. Mikheev AM, Mikheeva SA, Severs LJ, et al. Targeting TWIST1 through loss of function inhibits tumorigenicity of human glioblastoma. Mol Oncol. 2018;12(7):1188–1202. doi:10.1002/mol2.2018.12.issue-7
41. Pudova EA, Kudryavtseva AV, Fedorova MS, et al. HK3 overexpression associated with epithelial-mesenchymal transition in colorectal cancer. BMC Genomics. 2018;19(Suppl 3):113. doi:10.1186/s12864-018-4477-4
42. Zhou T, Li H, Zhong H, Zhong Z. Relationship between transforming growth factor-β1 and type 2 diabetic nephropathy risk in Chinese population. BMC Med Genet. 2018;19(1). doi:10.1186/s12881-018-0717-3
43. Nakano M, Kikushige Y, Miyawaki K, et al. Dedifferentiation process driven by TGF-beta signaling enhances stem cell properties in human colorectal cancer. Oncogene. 2019;38(6):780–793. doi:10.1038/s41388-018-0480-0
44. Zhang Y, Zhu M, Sun Y, Li W, Wang Y, Yu W. Upregulation of lncRNA casc2 suppresses cell proliferation and metastasis of breast cancer via inactivation of the TGF-β signaling pathway. Oncol Res. 2019;27(3):379–387. doi:10.3727/096504018X15199531937158
45. Wang L, Su N, Zhang Y, Wang G. Clinical significance of serum lncRNA cancer susceptibility candidate 2 (CASC2) for chronic renal failure in patients with Type 2 diabetes. Med Sci Monitor. 2018;24:6079–6084. doi:10.12659/MSM.909510
46. Barrow AD, Palarasah Y, Bugatti M, et al. OSCAR is a receptor for surfactant protein D that activates TNF-α release from human CCR2+ inflammatory monocytes. J Immunol. 2015;194(7):3317–3326. doi:10.4049/jimmunol.1402289
47. Schultz HS, Guo L, Keller P, et al. OSCAR-collagen signaling in monocytes plays a proinflammatory role and may contribute to the pathogenesis of rheumatoid arthritis. Eur J Immunol. 2016;46(4):952–963. doi:10.1002/eji.201545986
48. Takamura T, Honda M, Sakai Y, et al. Gene expression profiles in peripheral blood mononuclear cells reflect the pathophysiology of type 2 diabetes. Biochem Bioph Res Co. 2007;361(2):379–384. doi:10.1016/j.bbrc.2007.07.006
49. Irvine KM, Gallego P, An X, et al. Peripheral blood monocyte gene expression profile clinically stratifies patients with recent-onset type 1 diabetes. Diabetes. 2012;61(5):1281–1290. doi:10.2337/db11-1549
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.