Back to Journals » Cancer Management and Research » Volume 11
Identification of key genes and specific pathways potentially involved in androgen-independent, mitoxantrone-resistant prostate cancer
Authors Zhu S, Jiang LL, Wang LY, Wang LL, Zhang C, Ma Y, Huang T
Received 7 July 2018
Accepted for publication 16 November 2018
Published 3 January 2019 Volume 2019:11 Pages 419—430
DOI https://doi.org/10.2147/CMAR.S179467
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Rituraj Purohit
Sha Zhu,1 Lili Jiang,1,2 Liuyan Wang,3 Lingli Wang,1 Cong Zhang,1 Yu Ma,1 Tao Huang4
1Department of Immunology, Collaborative Innovation Center of Cancer Chemoprevention, College of Basic Medical Sciences, Zhengzhou University, Zhengzhou, Henan, China; 2Department of Basic Medicine, School of Nursing, Zhengzhou University, Zhengzhou, Henan, China; 3Department of Medicine, The Third People’s Hospital of Zhengzhou, Zhengzhou, Henan, China; 4Oncological Surgery, Cancer Hospital Affiliated to Zhengzhou University, Zhengzhou, Henan, China
Background: Resistance to mitoxantrone (MTX), an anthracenedione antineoplastic agent used in advanced and metastatic androgen-refractory prostate cancer (PCa), seriously limits therapeutic success.
Methods: Xenografts from two human PCa cell lines (VCaP and CWR22) were established in male severe combined immunodeficiency mice, and MTX was administered, with or without concurrent castration, three times a week until tumors relapsed. Microarray technology was used to screen for differentially expressed genes (DEGs) in androgen-independent, MTX-resistant PCa xenografts. Gene expression profiles of MTX-treatment xenografts and their respective parental cell lines were performed using an Agilent whole human genome oligonucleotide microarray and analyzed using Ingenuity Pathway Analysis software.
Results: A total of 636 genes were differentially expressed (fold change ≥1.5; P<0.05) in MTX-resistant castration-resistant prostate cancer (CRPC) xenografts. Of these, 18 were selected to be validated and showed that most of these genes exhibited a transcriptional profile similar to that seen in the microarray (Pearson’s r=0.87). Western blotting conducted with a subset of genes deregulated in MTX-resistant CRPC tumors was shown through network analysis to be involved in androgen synthesis, drug efflux, ATP synthesis, and vascularization.
Conclusion: The present data provide insight into the genetic alterations underlying MTX resistance in androgen-independent PCa and highlight potential targets to improve therapeutic outcomes.
Keywords: castration-resistant prostate cancer, gene expression profiling, drug resistance, differentially expressed genes
Introduction
Prostate cancer (PCa) is the most prevalent cancer and the second leading cause of cancer death among European and American men.1 Most patients presenting with advanced PCa have disease that is initially sensitive to androgen deprivation therapy (ADT), which successfully reduces tumor burden, improves symptoms, and can delay disease progression for several years.2,3 However, responses are not durable, and disease progression is inevitable. Mitoxantrone (MTX), a synthetic anthracenedione, has been routinely used for its palliative benefit and enhanced clinical remission alone or in combination with other drugs for the treatment of cancers.4–6 However, despite good initial response and survival benefits, most patients eventually develop resistance to this therapy.7
The anti-neoplastic activity of MTX is believed to be related to its ability to bind DNA and inhibit DNA topoisomerase II, an essential enzyme in DNA synthesis and cell division, which is highly expressed in cancer cells.8 Numerous changes in the genetic makeup of tumor cells occur typically during acquisition of drug resistance.9 The advent of microarray-based comparative genomic hybridization has greatly expanded our ability to identify genes involved in tumor chemotherapy resistance, to predict treatment response, and to forecast prognosis. In this study, the diversity and magnitude of transcriptional responses to genotoxic damage induced by MTX were assessed in two castration-resistant prostate cancer (CRPC) xenograft types and their parental cell lines using microarray-based gene expression profiling. Identification of differentially expressed genes (DEGs) in MTX-resistant CRPC may facilitate improved screening and further our understanding of the mechanisms mediating PCa relapse, paving the way for the development of targeted interventions that circumvent treatment resistance.
Materials and methods
Cell culture
The human VCaP and CWR22 PCa cell lines were obtained from Cell Bank of the Chinese Academy of Science (Shanghai, China). Cells were cultured in DMEM (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% FBS (HyClone, Logan, UT, USA) and 1% penicillin/streptomycin, at 37°C in a humidified atmosphere with 5% CO2. Confluent cells were harvested with 0.05% trypsin/0.53 mM EDTA, centrifuged 5 minutes at 1,200 rpm, and resuspended in complete medium at 1×107 cells/mL. Aliquots of 0.1 mL were used for subcutaneous injection into CB-17 severe combined immunodeficiency (SCID) mice (purchased from Guangzhou Provincial Medical Experimental Center).
Tumor inoculation and treatment
The animal study was carried out in a specific pathogen-free room and was approved by the medical ethics committee of the Zhengzhou University in accordance with the Guide for the Care and Use of Laboratory Animals (NIH publication no 80-23, revised 1996). Four- to six-week-old CB-17 male SCID mice were used in the experiments. Cells (1×106) were injected subcutaneously into both flanks resulting in two tumors per mouse. Once tumors became palpable, the mice were randomly divided into four treatment groups (six mice per group). In the first three groups, MTX was administered at 0.35, 1.0, and 3.5 mg/kg, respectively. The fourth group was treated with physiological saline (control) at the same time points. In another set of experiments, animals with palpable tumors were also assigned to one of four groups: MTX (3.5 mg/kg), castration, MTX (3.5 mg/kg) in combination with castration, and control. Surgical castration was performed after tumors had developed. In all experiments, MTX and saline were administered intragastrically in a 100 µL volume three times a week. Tumor diameters were measured with a caliper twice a week until the animals were sacrificed after 6 weeks of treatment. Tumor weight was calculated by the formula: tumor weight (mg) = (length×width2)/2.
RNA processing and microarray scanning
Total tumor RNA was extracted using Trizol reagent (Takara, Dalian, China), and concentrations determined by a spectrophotometer (NanoDrop; Nyxor Biotech, Paris, France). All the processes were carried out according to the manufacturers’ instructions. Enrichment of total RNA from samples was carried out using the RNeasy Micro kit (Qiagen NV, Venlo, the Netherlands), and samples’ quality and quantity were assessed on a spectrophotometer. Briefly, 1 µg of total RNA from each sample was amplified and transcribed into fluorescent cRNA using the protocol provided with Agilent’s Quick Amp Labeling kit. Labeled cRNAs were hybridized onto the Whole Human Genome Oligo Microarray (4×44K; Agilent Technologies, Santa Clara, CA, USA) in Agilent’s SureHyb Hybridization Chambers. Sample preparation and microarray hybridization were performed based on the manufacturer’s standard protocols, and the microarray procedure conformed to quality standards. Washes and scanning of the arrays were carried out according to manufacturer’s instructions. Images were autogridded, and the chemiluminescent signals were quantified, corrected for background at each spot, and spatially normalized. DEGs were identified through filtering the dataset using P<0.01 and a signal-to-noise ratio >2 for use in ANOVA statistical analysis. Three representative xenografts were chosen from each treatment cohort for the microarray experiments, and data are shown as mean values.
Gene expression data validation by quantitative reverse transcription PCR (qRT-PCR)
qRT-PCR was performed with QPK-201 SYBR Green master mix (Toyobo, Osaka, Japan) and the ABI 7300 system from Thermo Fisher Scientific. The primers used in the study were obtained from Thermo Fisher Scientific. Thermocycling parameters included an room temperature step at 50°C for 20 minutes, followed by a DNA polymerase activation step at 95°C for 2 minutes and 50 PCR cycles (95°C for 20 seconds and 60°C for 30 seconds). All reactions were conducted in triplicate. Differential expression (fold change) for each gene was calculated using the comparative CT method.
Pathway enrichment and network construction
The GeneSpring GX software package (Agilent Technologies) was used to statistically analyze 636 DEGs. To assess the relationships between DEGs and signaling pathways, additional filtering (minimum three-fold change) was applied to extract the most relevant DEGs, which were analyzed using Ingenuity Pathway Analysis (IPA) software. Genes with known gene symbols and their corresponding expression values were uploaded into the software. Each gene symbol was mapped to its corresponding gene object in the Ingenuity Knowledge Base. Networks of these genes were algorithmically generated based on their connectivity.
Western blot analysis
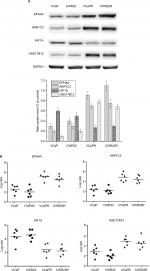
Protein samples from total cell lysates were separated on 10% polyacrylamide resolving gels and transferred onto nitrocellulose membranes for 2 hours at 250 mA. Protein binding sites on membranes were blocked for 1 hour at 25°C in 5% (w/v) Marvel’s dried skimmed milk/PBS/3% (v/v) Tween-20 (PBST), then incubated overnight at 4°C with EPHA4, AKR1C3, HIF1A, and HSD17B12 monoclonal antibodies (1:1,000 dilution; Thermo Fisher Scientific). The membranes were washed 3×10 minutes in tris buffered saline tween and probed with horseradish peroxidase-conjugated secondary antibodies (Amersham Life Sciences, Buckinghamshire, UK) for 1 hour at 25°C. Following 3×10-minute washes in PBST, bands were detected using enhanced chemiluminescence (ECL+ reagents; Amersham Life Sciences). Densitometric quantification of band intensities was performed using Kodak one-dimensional image analysis software.
Statistical analyses
Results are presented as mean ± standard error of the mean. The Wilcoxon rank-sum test of variance was performed to determine whether differences between groups were statistically significant, with P<0.05 set as threshold to indicate significance. SPSS 12.0 software was used for statistical analyses.
Results
MTX and androgen sensitivity testing
To investigate the sensitivity to MTX of human PCa cells in vivo, 106 PCa cells (from VCaP or CWR22 cell lines) were inoculated into the flanks of 5-week-old male CB-17 SCID mice to generate subcutaneous tumors. In general, palpable tumors developed 4–5 weeks after implantation. Once tumors grew to nearly 200 mm3, mice were treated intragastrically with different concentrations of MTX. Tumor volume and weight were measured twice weekly. As shown in Figure 1A and B, tumor volumes differed significantly between control and MTX-treated mice (P<0.01). MTX decreased tumor growth in a dose-dependent manner; although no significant effect was noted with the lowest dose, a 38.2% and 34.3% volume reduction was observed with 3.5 mg/kg MTX in VCaP and CWR22 xenografts, respectively. However, all mice eventually relapsed and tumors became resistant to MTX.
SCID mice with palpable PCa xenografts were castrated and treated with 3.5 mg/kg MTX to evaluate potential synergistic antitumor activities of androgen deprivation and MTX. This showed that VCaP and CWR22 were androgen dependent, and surgical castration led to a statistically significant (P<0.05) decrease in the tumor size of VCaP and CWR22 xenografts (Figure 1A and B). Although both VCaP and CWR22 xenografts are castration sensitive, these xenografts eventually developed to androgen-independent phenotype associated with regrowth. It demonstrated that tumor growth was significantly inhibited in castrated mice concurrently treated with MTX (P<0.01). As shown in Figure 1C and D, tumors of VCaP and CWR22 in the combination group were significantly smaller than those in the control group (P<0.01), the MTX-only group (P<0.05), and the castration-only group (P<0.05). Although significant reductions in median tumor size were recorded for the combination group on days 24, 35, 42, and 51, treatment resistance, evidenced by tumor regrowth, developed thereafter. All mice consistently maintained their body weight during each study.
Microarray-based identification of DEGs in MTX-resistant CRPC xenografts
Identification of DEGs between MTX-treatment xenografts and their parent PCa cell lines was carried out by microarray analysis on an Agilent Whole Human Genome Oligo Microarray platform. Comparative analysis was done after applying a strict threshold. We found that 636 genes were differentially expressed (P<0.05) in MTX-resistant CRPC xenografts. From these, we selected the top 20 upregulated and downregulated genes according to the log ratio expression values (Tables 1 and 2), and among these, the ones in the MTX-resistant CRPC groups whose expression levels were changed more than three fold (P<0.01) were compared with their parent groups. Upon comparison of DEGs from both xenografts (VCaPR and CWR22R), we found that nearly a quarter of the genes overlap.
  | Table 1 Top ten up- and downexpressed genes in xenograft of VCaP vs its parent cells |
  | Table 2 Top ten up- and downexpressed genes in xenograft of CWR22R vs its parent cells |
Validation of gene expression data by qRT-PCR and Western blotting
The expression patterns of four DEGs, EPHA4, AKR1C3, HIF1A, and HSD17B12, were evaluated by Western blot (Figure 2A) and qRT-PCR (Figure 2B). Results confirmed upregulated EPHA4, AKR1C3, and HSD17B12 expression in both VCaPR and CWR22R MTX-resistant CRPC xenografts. In contrast, HIF1A expression was markedly downregulated in both tumor types, compared with their respective parental cell lines. In addition, a panel of 18 DEGs with the highest and lowest expression differences between both VCaPR and CWR22R MTX-treatment xenografts and their parental cells lines was selected and tested by qRT-PCR (Figure 3A and B). Results showed that most of these genes exhibited a transcriptional profile similar to that seen in the microarray (Pearson’s correlation coefficient=0.87). Hence, the microarray provided a reliable assessment of gene expression differences between MTX-treatment xenografts and their originating PCa cell lines.
Canonical pathways and network analysis
By considering only probe sets common to both MTX-resistant PCa xenografts, 252 genes were qualified as network and function eligible by IPA. The DEGs identified in the two sets of data were then analyzed by the IPA library of canonical pathways. The ten most significant canonical pathways affected by these DEGs were mainly related to DNA replication, recombination, protein expression, and cell cycle (Figure 4A). Four pathways, namely PCa signaling, PI3K/AKT signaling, glioblastoma multiforme signaling, and PTEN signaling, involved in cellular growth, proliferation, and development, showed significant enrichment.
We further identified potential biological networks likely affected by these DEGs to better define how individual DEGs interact in specific pathways. Specifically, network 1 (Figure 4B) was centered on the T3-TR-RXR pathway, in which the thyroid hormone receptor (TR) and the retinoid X receptor (RXR) act as heterodimers (TR-RXR) that preferentially bind thyroid hormone T3, activating gene transcription through coactivator recruitment. These gene products indirectly interact with an important nuclear receptor, NR1H4, for cell survival, proliferation, and upregulation of enzymes, including CYP17A1, AKR1C3, and AKR1C1/AKR1C2 involved in androgen synthesis. Network 2 (Figure 4C) is centered on the NF-κB pathway, associated with cell proliferation, differentiation, and DNA damage response. Here, several members of the UDP-glucuronosyltransferase 2B (UGT2B) gene family, including UGT2B4, UGT2B10, UGT2B7, and UGT2B28, were highly upregulated. The UGT2B gene subfamily is involved in the metabolic clearance of numerous endogenous compounds, including bile acids and steroid hormones, as well as exogenous agents including various carcinogens and drugs.10,11 Apoptosis-related genes such as apoptosis 9, apoptosis 10, CYP27B1, and GPX1 were significantly downregulated in MTX-treatment xenografts and are also shown in the network. These DEGs are mainly associated with regulation of proliferation, gene expression, differentiation, and cell survival.
Discussion
Many molecular methods have been used in the attempt to elucidate the heterogeneous nature of PCa, determine the mechanisms behind its development, and propose new therapeutic and prognostic targets.12 Gene expression profiling or genome scale analysis has proven to be successful in various experimental settings and has the potential to highlight the dynamic molecular diversity encountered during tumor progression.13 Since studies investigating gene expression changes during progression of MTX-resistant CRPC are lacking, this study sought to identify DEGs in MTX refractory CRPC that may be exploited as specific prognostic and therapeutic targets.
Microarray and bioinformatics analyses revealed overexpression of fractalkine, also known as chemokine (C-X3-C motif) ligand 1 (CX3CL1), in VCaPR and CWR22R xenografts. CX3CL1 is the only member recognized so far of the CX3C chemokine subfamily and was reported to participate in the regulation of cell adhesion, migration, and survival in human PCa cells by interacting with its receptor CX3CR1, primarily expressed on macrophages, circulating monocytes, and natural killer cells.14,15 Importantly, CX3CL1-CX3CR1 binding plays a crucial role in epithelial-to-mesenchymal transition (EMT), PCa progression, and skeletal metastasis,16 and was proposed to inhibit the apoptosis of cancer cells through MAPK/ERK activation.17
The EPHA4 gene was also found to be highly upregulated in MTX-resistant CRPC xenografts. This gene belongs to the ephrin receptor subfamily of the protein-tyrosine kinase family. Ephrin ligands and receptors are differentially expressed on arteries and veins during development.18 Altered expression patterns of EPHA4/ephrins have been correlated with tumor invasiveness, vascularization, and metastatic potential.19 Moreover, high levels of EPHA4 mRNA correlate significantly with reduced overall survival in cancer patients.20
We also observed overexpression of ABCG2, the subfamily G of the large human ATP-binding cassette (ABC) transporter superfamily, in MTX-resistant VCaPR and CWR22R CRPC xenografts. ABCG2 forms homodimers or heterodimers with other members of the ABCG subfamily to function as an efflux transporter.21 ABCG2 is highly expressed in prostate stem cells and plays an important role in regulating intracellular androgen levels by mediating androgen efflux.22 Importantly, ABCG2 is also considered the main contributing factor to drug resistance in ovarian carcinoma xenografts.23 ABCG2 requires cellular ATP for transporting its substrates.24 Cytochrome c oxidase (COX) is a component of the respiratory chain complex involved in oxidative phosphorylation and ATP production.25 Accordingly, the upregulation of COX6c found in MTX-resistant VCaPR and CWR22R tumors might contribute to supply the large amount of ATP required by ABCG2 to pump MTX out of the cells.
Deregulated AKR1C3 expression has been associated with squamous cell carcinoma of the head and neck and other human cancers.26,27 The AKR enzymes comprise a functionally diverse gene family, with four AKR1C isoforms (AKR1C1–4) presently identified.28 Although low levels of AKR1C3 and AKR1C1 were observed in normal prostatic epithelium,29 this study showed significant upregulation of AKR1C3 in MTX-resistant PCa xenografts. Knuuttila et al30 suggested that castration induces the upregulation of AKR1C3 and other enzymes, such as CYP17A1 and HSD17B6, involved in estrogen and androgen metabolism; activates insulin-like growth factor (IGF)-1 and Akt signaling; and promotes tumor angiogenesis and aggressiveness. In addition, AKR1C3 may also promote CRPC progression by activating 17β-estradiol-mediated signaling pathways.31
Network analysis helped us obtain global and integrated molecular information about interactions between MTX-related DEGs. One important gene network was identified around the NF-κB gene. NF-κB is found in almost all animal cell types and is involved in cellular responses to stimuli such as stress, cytokines, free radicals, and ultraviolet irradiation.32–34 Deregulation of NF-κB signaling has been linked to cancer and inflammatory and autoimmune diseases.35,36 In relation to this network, we observed significant downregulation of the pro-apoptotic caspase-10 gene in MTX-treatment xenografts. Study indicated that caspase-10 mRNA expression decreased significantly in stage II colorectal cancer tissues that predict poor prognosis in patients.37 Dysregulation of CYP27B1 was also observed in the NF-κB signaling network. A study indicated that repression of CYP27B1 gene expression, mediated by interaction of GFI1 with other proteins to form a large protein complex, leads to cancer progression.38 Another study demonstrated that mRNA expression of CYP27B1 in the malignant breast tissue was lower, compared with that in the normal tissue and suggested that evaluating the mRNA expression levels of CYP27B1 may be useful for estimating the vitamin D anabolism and catabolism and risk of the progression of breast cancer. The carcinogenesis-related gene BCL11A, a DNA sequence-specific transcriptional repressor, was instead upregulated and interacted directly with NF-κB in the network. Overexpression of BCL11A may play a primary role in the pathogenesis of lymphoid malignancies,39 is correlated with advanced clinical N1/N2 lymphatic metastasis in patients,40 and was shown to be a prognostic factor for both overall survival and disease-free survival.41,42 In this study, BCL11A expression was dramatically higher in the MTX-treatment xenografts than in their parental cell lines; BCL11A was also overexpressed and play a positive role in several other types of human cancers such as NSCLC, breast cancer, squamous cell carcinoma, and large cell carcinoma.43–45
Conclusion
Since some gene interactions may be specific of different cellular/experimental conditions, clinical validation of the present results is warranted. Nevertheless, this exploratory analysis may be useful to bestow a theranostic perspective to the current trend of research in PCa and to help identify molecular targets to overcome castration and MTX chemotherapy resistance.
Data availability
The data of the study have been deposited into the Research Data Deposit (http://www.researchdata.org.cn with the Approval Number asRDDB2018000383).
Acknowledgment
This study was supported by grants from Medical Key Technologies R&D Program of Henan Province (201402004) and Natural Science and Technologies Program of Henan Province (172102410018).
Author contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24(14):2137–2150. | ||
Rydzewska LHM, Burdett S, Vale CL, et al. Adding abiraterone to androgen deprivation therapy in men with metastatic hormone-sensitive prostate cancer: a systematic review and meta-analysis. Eur J Cancer. 2017;84:88–101. | ||
Li Y, Li CX, Ye H, et al. Decrease in stromal androgen receptor associates with androgen-independent disease and promotes prostate cancer cell proliferation and invasion. J Cell Mol Med. 2008;12(6B):2790–2798. | ||
Nair RV, Santhakumar H, Jayasree RS. Gold nanorods decorated with a cancer drug for multimodal imaging and therapy. Faraday Discuss. 2018;207:423–435. | ||
Wang AY, Weiner H, Green M, et al. A phase I study of selinexor in combination with high-dose cytarabine and mitoxantrone for remission induction in patients with acute myeloid leukemia. J Hematol Oncol. 2018;11(1):4. | ||
Aguirre-Hernández C, Maya-Pineda H, Millán JS, et al. Sensitisation to mitoxantrone-induced apoptosis by the oncolytic adenovirus Ad∆∆ through Bcl-2-dependent attenuation of autophagy. Oncogenesis. 2018;7(1):6. | ||
Flaig TW, Potluri RC, Ng Y, Todd MB, Mehra M. Treatment evolution for metastatic castration-resistant prostate cancer with recent introduction of novel agents: retrospective analysis of real-world data. Cancer Med. 2016;5(2):182–191. | ||
Salerno S, da Settimo F, Taliani S, et al. Recent advances in the development of dual topoisomerase I and II inhibitors as anticancer drugs. Curr Med Chem. 2010;17(35):4270–4290. | ||
Gomez-Sarosi L, Sun Y, Coleman I, Bianchi-Frias D, Nelson PS. DNA Damage Induces a Secretory Program in the Quiescent TME That Fosters Adverse Cancer Phenotypes. Mol Cancer Res. 2017;15(7):842–851. | ||
Collier AC, Ganley NA, Tingle MD, et al. UDP-glucuronosyltransferase activity, expression and cellular localization in human placenta at term. Biochem Pharmacol. 2002;63(3):409–419. | ||
Hu DG, Mackenzie PI, Mckinnon RA, Meech R. Genetic polymorphisms of human UDP-glucuronosyltransferase (UGT) genes and cancer risk. Drug Metab Rev. 2016;48(1):47–69. | ||
Guo X, Zhang C, Guo Q, et al. The homogeneous and heterogeneous risk factors for the morbidity and prognosis of bone metastasis in patients with prostate cancer. Cancer Manag Res. 2018;10:1639–1646. | ||
Jamieson NB, Maker AV. Gene-expression profiling to predict responsiveness to immunotherapy. Cancer Gene Ther. 2017;24(3):134–140. | ||
Szukiewicz D, Kochanowski J, Mittal TK, Pyzlak M, Szewczyk G, Cendrowski K. CX3CL1 (fractalkine) and TNFα production by perfused human placental lobules under normoxic and hypoxic conditions in vitro: the importance of CX3CR1 signaling. Inflamm Res. 2014;63(3):179–189. | ||
Park MH, Lee JS, Yoon JH. High expression of CX3CL1 by tumor cells correlates with a good prognosis and increased tumor-infiltrating CD8+ T cells, natural killer cells, and dendritic cells in breast carcinoma. J Surg Oncol. 2012;106(4):386–392. | ||
Borsig L, Wolf MJ, Roblek M, Lorentzen A, Heikenwalder M. Inflammatory chemokines and metastasis-tracing the accessory. Oncogene. 2014;33(25):3217–3224. | ||
Zhang W, Thompson BJ, Hietakangas V, Cohen SM. MAPK/ERK signaling regulates insulin sensitivity to control glucose metabolism in Drosophila. PLoS Genet. 2011;7(12):e1002429. | ||
Goldshmit Y, Spanevello MD, Tajouri S, et al. EphA4 blockers promote axonal regeneration and functional recovery following spinal cord injury in mice. PLoS One. 2011;6(9):e24636. | ||
Husa AM, Magic´ Ž, Larsson M, Fornander T, Pérez-Tenorio G. EPH/ephrin profile and EPHB2 expression predicts patient survival in breast cancer. Oncotarget. 2016;7(16):21362–22180. | ||
Oshima T, Akaike M, Yoshihara K, et al. Overexpression of EphA4 gene and reduced expression of EphB2 gene correlates with liver metastasis in colorectal cancer. Int J Oncol. 2008;33(3):573–577. | ||
Kowalski P, Stein U, Scheffer GL, Lage H. Modulation of the atypical multidrug-resistant phenotype by a hammerhead ribozyme directed against the ABC transporter BCRP/MXR/ABCG2. Cancer Gene Ther. 2002;9(7):579–586. | ||
Gangavarapu KJ, Azabdaftari G, Morrison CD, et al. Aldehyde dehydrogenase and ATP binding cassette transporter G2 (ABCG2) functional assays isolate different populations of prostate stem cells where ABCG2 function selects for cells with increased stem cell activity. Stem Cell Res Ther. 2013;4(5):132. | ||
Ricci JW, Lovato DM, Severns V, Sklar LA, Larson RS. Novel ABCG2 Antagonists Reverse Topotecan-Mediated Chemotherapeutic Resistance in Ovarian Carcinoma Xenografts. Mol Cancer Ther. 2016;15(12):2853–2862. | ||
Sobek KM, Cummings JL, Bacich DJ, O’Keefe DS. Contrasting roles of the ABCG2 Q141K variant in prostate cancer. Exp Cell Res. 2017;354(1):40–47. | ||
Bauerfeld CP, Rastogi R, Pirockinaite G, et al. TLR4-mediated AKT activation is MyD88/TRIF dependent and critical for induction of oxidative phosphorylation and mitochondrial transcription factor A in murine macrophages. J Immunol. 2012;188(6):2847–2857. | ||
Miller VL, Lin HK, Murugan P, et al. Aldo-keto reductase family 1 member C3 (AKR1C3) is expressed in adenocarcinoma and squamous cell carcinoma but not small cell carcinoma. Int J Clin Exp Pathol. 2012;5(4):278–289. | ||
O’Reilly MW, Kempegowda P, Walsh M, et al. AKR1C3-Mediated Adipose Androgen Generation Drives Lipotoxicity in Women with Polycystic Ovary Syndrome. J Clin Endocrinol Metab. 2017;102(9):3327–3339. | ||
Davies NJ, Hayden RE, Simpson PJ, et al. AKR1C isoforms represent a novel cellular target for jasmonates alongside their mitochondrial-mediated effects. Cancer Res. 2009;69(11):4769–4775. | ||
Karunasinghe N, Masters J, Flanagan JU, Ferguson LR. Influence of Aldo-keto Reductase 1C3 in Prostate Cancer – A Mini Review. Curr Cancer Drug Targets. 2017;17(7):603–616. | ||
Knuuttila M, Yatkin E, Kallio J, et al. Castration induces up-regulation of intratumoral androgen biosynthesis and androgen receptor expression in an orthotopic VCaP human prostate cancer xenograft model. Am J Pathol. 2014;184(8):2163–2173. | ||
Fankhauser M, Tan Y, Macintyre G, et al. Canonical androstenedione reduction is the predominant source of signaling androgens in hormone-refractory prostate cancer. Clin Cancer Res. 2014;20(21):5547–5557. | ||
Brasier AR. The NF-kappaB regulatory network. Cardiovasc Toxicol. 2006;6(2):111–130. | ||
Bakhoum SF, Ngo B, Laughney AM, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature. 2018;553(7689):467–472. | ||
Park SH, Cho JY, Oh SW, et al. Arctigenin protects against ultraviolet-A-induced damage to stemness through inhibition of the NF-κB/MAPK pathway. Chem Biol Interact. 2018;282:63–68. | ||
Ma J, Sun X, Guo T, et al. Interleukin-1 receptor antagonist inhibits angiogenesis via blockage IL-1α/PI3K/NF-κβ pathway in human colon cancer cell. Cancer Manag Res. 2017;9:481–493. | ||
Lin G, Li C, Huang C. Co-expression of NF-κB-p65 and phosphorylated NF-κB-p105 is associated with poor prognosis in surgically resectable non-small cell lung cancer. J Cell Mol Med. 2018; 22(3):1923–1930. | ||
Shen XG, Wang C, Li Y, et al. Downregulation of caspase-10 predicting poor survival after resection of stage II colorectal cancer. Int J Colorectal Dis. 2011;26(12):1519–1524. | ||
Dwivedi PP, Anderson PH, Tilley WD, May BK, Morris HA. Role of oncoprotein growth factor independent-1 (GFI1) in repression of 25-hydroxyvitamin D 1alpha-hydroxylase (CYP27B1): a comparative analysis in human prostate cancer and kidney cells. J Steroid Biochem Mol Biol. 2007;103(3-5):742–746. | ||
Satterwhite E, Sonoki T, Willis TG, et al. The BCL11 gene family: involvement of BCL11A in lymphoid malignancies. Blood. 2001;98(12):3413–3420. | ||
Yin J, Zhang F, Tao H, et al. BCL11A expression in acute phase chronic myeloid leukemia. Leuk Res. 2016;47:88–92. | ||
Jiang BY, Zhang XC, Su J, et al. BCL11A overexpression predicts survival and relapse in non-small cell lung cancer and is modulated by microRNA-30a and gene amplification. Mol Cancer. 2013;12:61. | ||
Khaled WT, Choon Lee S, Stingl J, et al. BCL11A is a triple-negative breast cancer gene with critical functions in stem and progenitor cells. Nat Commun. 2015;6:5987. | ||
Zhang N, Jiang BY, Zhang XC, et al. The BCL11A-XL expression predicts relapse in squamous cell carcinoma and large cell carcinoma. J Thorac Dis. 2015;7(9):1630–1636. | ||
Santuario-Facio SK, Cardona-Huerta S, Perez-Paramo YX, et al. A New Gene Expression Signature for Triple Negative Breast Cancer Using Frozen Fresh Tissue before Neoadjuvant Chemotherapy. Mol Med. 2017;23:1. | ||
Jamal-Hanjani M, Wilson GA, Mcgranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med. 2017;376(22):2109–2121. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.