Back to Journals » International Journal of General Medicine » Volume 15
Identification of Hub Genes and Biological Pathways in Inclusion Body Myositis Using Bioinformatics Analysis
Authors Wu Y, Zhao Z , Zhang J, Wang Y, Song X
Received 24 November 2021
Accepted for publication 12 January 2022
Published 9 February 2022 Volume 2022:15 Pages 1281—1293
DOI https://doi.org/10.2147/IJGM.S346965
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Yue Wu,1,2,* Zijun Zhao,3,* Jinru Zhang,1,2 Yaye Wang,1,2 Xueqin Song1,2
1Department of Neurology, The Second Hospital of Hebei Medical University, Shijiazhuang, Hebei, 050000, People’s Republic of China; 2Neurological Laboratory of Hebei Province, Shijiazhuang, Hebei, People’s Republic of China; 3Department of Neurosurgery, The Second Hospital of Hebei Medical University, Shijiazhuang, Heibei, 050000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xueqin Song
Department of Neurology, The Second Hospital of Hebei Medical University, 215 Heping West Road, Shijiazhuang, Hebei, 050000, People’s Republic of China
, Tel/Fax +86-318-2187209
, Email [email protected]
Background: Inclusion body myositis (IBM) is a unique idiopathic inflammatory myopathy with unclear pathogenesis and poor prognosis. Although previous publications have identified some molecular biomarkers, the value of these biomarkers is unknown.
Objective: To identify hub genes and signaling pathways related to IBM for understanding the IBM-related mechanisms and providing guidance for therapy development.
Methods: Two microarray datasets (GSE3112 and GSE128470) were downloaded from the Gene Expression Omnibus (GEO) database. GEO2R was used to detect differentially expressed genes (DEGs) between IBM and normal muscle tissues. The hub genes were determined using protein–protein interaction (PPI) network in Cytoscape. The specific signaling pathways and biological functions of IBM were identified using GO, KEGG, and GSEA enrichment analyses. Moreover, CIBERSORT was applied to estimate the expression level of 22 immune cell types in IBM and normal muscle tissue. The relationship between the immune cell types and hub genes was then explored.
Results: A total of 219 DEGs and 10 hub genes were identified. Enrichment analyses revealed that the chemokine signaling pathway, cellular response to interferon-gamma, and P53 pathway have crucial roles in IBM. Immune infiltration analyses showed that IBM was associated with high level of CD8 T cells, Tregs, and macrophages. Finally, five potential drugs were predicted for IBM patients through CMap (connectivity map) database.
Conclusion: In this study, the underlying molecular mechanisms and immunological landscape of IBM were investigated, and thus may provide new directions for future research on IBM pathogenesis.
Keywords: bioinformatic analysis, inclusion body myositis, differentially expressed genes, biological pathways, immune infiltrating cell
Introduction
Inclusion body myositis (IBM) is an acquired inflammatory myopathy. Disease onset is gradual and usually begins after the age of 50 years.1 IBM is characterized by asymmetrical muscular weakness and atrophy, mostly affecting quadriceps femoris and deep finger flexors.2 IBM is a rare sporadic disease with an average prevalence of 24.8–45.6/1,000,000.1,3 IBM has late-onset, asymmetric muscle weakness and progresses slowly, compared with other inflammatory myopathies, such as dermatomyositis and polymyositis. Besides inflammatory infiltrates, the muscle pathology of IBM is associated with rimmed vacuoles and protein aggregation. Unlike patients with other inflammatory myopathy, glucocorticoid, azathioprine, methotrexate, and other immunosuppressants or immunomodulators rarely benefit IBM patients.4 Moreover, there is no effective treatment for IBM patients due to the unclear etiology and pathogenesis. Previous studies have demonstrated that inflammatory and degenerative pathways played important roles in IBM.2 Inflammatory infiltrate, abnormal protein deposition, and mitochondrial dysfunction occur in the muscle of IBM patients. Therefore, the precise molecular mechanisms and IBM pathways should be assessed to develop effective IBM treatment strategies.
Tremendous advances have been achieved in the genetic analysis of IBM in recent years due to the development of chip techniques and sequencing approaches. The international Myositis Genetics Consortium (MYOGEN) assessed 252 IBM patients and showed that HLA-DRB1*03:01(p = 5.77×10−34), HLA-DRB1*01:01(p = 1.57×10−16), and HLA-DRB1*13:01(p = 3.8×10−8) are associated with a higher risk of IBM.5 Although HLA-DRB1*03:01 is associated with dermatomyositis, polymyositis and polymyositis, it is not the specific risk factor for IBM. Therefore, it is necessary to identify other indicators for the diagnosis and prognostic assessment of IBM patients.
Microarray technology has been widely used to study various diseases since it can simultaneously monitor several gene expression profiles. However, it is difficult to identify reliable key genes and pathways using microarray technology due to the false-positive rates of independent microarray studies. Herein, two microarray datasets of healthy and IBM samples, GSE3112 and GSE128470 were downloaded from the Gene Expression Omnibus (GEO) database. Bioinformatics methods were used to identify differentially expressed genes (DEGs). The DEGs were subsequently analyzed using PPI (protein-protein interaction), GO (Gene Ontology), and KEGG (Kyoto Encyclopaedia of Genes and Genomes). This research provides a preliminary understanding of the molecular pathogenesis of IBM, and several potential therapeutic targets for IBM treatment.
Materials and Methods
Microarray Data
The publicly available microarray data were obtained from GSE3112 and GSE128470 in GEO (ncbi.nlm.nih.gov/geo/). The expression profile GSE3112 had 23 IBM samples and 11 healthy samples, while the GSE128470 profile contained 12 healthy samples, 26 IBM samples, 12 DM (dermatomyositis) samples, six NM (necrotizing myopathy) samples, 14 NS (non-specific myositis) samples, and seven PM (polymyositis) samples. The sample information of IBMs and normal muscle tissues in the two datasets were showed in Table S1 (GSE3112) and Table S2 (GSE128470).
Identification of DEGs and Enrichment Analysis
GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/) was used to identify DEGs between normal muscle tissues and IBM. The screening criteria for DEG determination were: | log2 fold change (log2FC) | ≥1.5 and P-value <0.5. The intersection of DEGs in the two datasets was conducted for further investigation. KEGG, GO, and GSEA (Gene set enrichment analysis) enrichment were performed using clusterProfiler R package to explore the functions of the DEGs.
Protein-Protein Interaction (PPI) Network and Subcluster Analysis
The protein-protein interactions of DEGs were predicted using the STRING online platform (http://string-db.org/). Molecular Complex Detection (MCODE) within Cytoscape was used to explore the gene clusters in the PPI network.6
Expression and ROC Analysis
The expression profiles of IBM and healthy muscle tissues were used to identify 10 DEGs with the highest scores. The Receiver operating characteristic (ROC) curves were generated using pROC R package. The ROC curves were used to evaluate the predictive power of the identified biomarkers.
Sample Acquisition and Immunohistochemistry
One normal muscle tissue and three IBM muscle tissues were obtained from the Second Hospital of Hebei Medical University, which was approved by Research Ethics Committee of the Second Hospital of Hebei Medical University. IBM diagnosis was conducted based on the diagnostic clinicopathological features.
The muscle samples were frozen, 5-μm-thick cryosections were cut under −20 °C. A citrate buffer (pH = 6.0) was used for antigen retrieval. Then rinsed twice using PBS for 5 minutes each time. The sections were then incubated with 10% goat serum at room temperature for 30 minutes to block non-specific binding sites. The goat serum was removed, then the samples were incubated with CD4 (Abcam ab183685), ITGAM (Abcam ab287572), and CCR5 (Abcam ab287959) at 4 °C overnight. The samples were then washed thrice (3 minutes each) using PBS. The samples were then incubated with a secondary antibody at 37 °C for 1 h. The samples were washed thrice with PBS, then each section was stained with DAB for visualization using the microscope.
Immune Infiltration Analysis
The deconvolution approach CIBERSORT has been widely used to study various diseases, including ulcerative colitis,7 lupus,8 and rheumatoid arthritis, due to its validity and reliability.9 The white blood cell gene bio-signature matrix in CIBERSORT comprises 547 genes, known as LM22, which is used to differentiate the 22 immune cell types consisting of regulatory T cells (Tregs), naive CD4 T cells, macrophages M2, activated memory CD4 T cells, plasma cells, follicular helper T cells, gamma delta T cells, CD8 T cells, macrophages M1, memory B cells, activated NK cells, macrophages M0, resting NK cells, monocytes, resting mast cells, resting memory CD4 T cells, activated mast cells, resting dendritic cells, neutrophils, naive B cells, activated dendritic cells, and eosinophils.10 Herein, CIBERSORT was used to determine the proportions of the 22 immune cells in the IBM and normal samples of GSE3112.
Gene Set Variation and Enrichment Analysis
GSVA (Gene set variation analysis) is a non-parametric and unsupervised tool used to estimate certain pathways or biological functions via the sample expression data.11 Herein GSVA analysis was performed using “h.all.v7.2.symbols” gene sets (obtained from the MSigDB (Molecular Signatures Databases)) to explore the differences in biological processes between IBM and normal muscle tissues. Gene set enrichment analysis (GSEA) was used to validate the characteristics and the potential activated pathways of IBM in GSVA results.
Connectivity Map Analysis
Connectivity map (CMap)12 was used to explore potential drugs for IBM. Enrichment score (>0.8) and P-value (< 0.01) were considered the screening standard.
Results
DEGs Identification
A total of 466 DEGs were obtained from GSE3112 (nine down-regulated and 457 up-regulated) (Figure 1A), and 310 DEGs were obtained from GSE128470 (23 down-regulated and 287 up-regulated) (Figure 1B). The DEGs could clearly distinguish between healthy and IBM samples, as shown in the heatmaps obtained through cluster analysis (Figure 1D and E). A Venn diagram identified 219 overlapping genes between GSE3112 and GSE128470, which were used for further analysis (Figure 1C).
PPI Network and Pathway Enrichment Analysis
The PPI analysis results conducted using STRING tools are shown (Figure 2A). The MCODE method identified four subclusters for the PPI networks (Figure 2B–E). GO analysis showed that the DEGs were mainly enriched in leukocyte migration, regulation of immune effector process, endocytic vesicle membrane, external side of the plasma membrane, chemokine receptor binding, and peptide antigen binding (Figure 2F). KEGG analysis showed that the DEGs were significantly enriched in phagosome, chemokine, Natural killer cell-mediated cytotoxicity, and Th1, Th2, and Th17 cell differentiation signaling pathways (Figure 2G).
Moreover, CD4 (degree=79), PTPRC (degree=70), ITGAM (degree=58), TYROBP (degree=55), CCR5 (degree=53), IRF8 (degree=51), STAT1 (degree=50), ITGB2 (degree=49), HLA-DRA (degree=47), and CXCL10 (degree=47) were screened as hub genes of IBM.
Differential Gene Expression and ROC Analyses
In this study, the GSE3112 and GSE128470 gene data sets were used to detect the 10 hub genes The hub genes were significantly up-regulated in IBM (P<0.05) (Figure 3A and B). The ROC curves demonstrated that these hub genes are potential biomarkers for IBM diagnosis (Figure 3C and D). Further analysis suggested that the hub genes were positively correlated with each other (Figure 3E and F).
The expression level of the hub genes in IBM and other idiopathic inflammatory myopathies, including dermatomyositis (DM), necrotizing myopathy (NM), nonspecific myositis (NS), and polymyositis (PM) were then determined based on the GSE128470 cohort. The expression of several hub genes (CCR5, HLA-DRA, and PTPRC) could distinguish IBM and DM, NM, NS. Moreover, the expression of the hub genes was not significantly different between IBM and PM (Figure 4).
 |
Figure 4 Expression level of the 10 hub genes in IBM and other idiopathic inflammatory myopathies ns: no significance; *P < 0.05; **P <0.01; ***P < 0.001. |
Immunohistochemistry
Previous studies have reported that CD4 and CCR5 play an important role in IBM.13 ITGAM is associated with the inflammatory response.14 Immunohistochemistry showed that CD4, CCR5, and ITGAM were highly expressed in IBM than in normal muscle tissues (Figure 5). It was in keeping with the results of the biomarker analysis.
 |
Figure 5 Immunohistochemistry Immunohistology revealed the expression of CD4, CCR5, and ITGAM in IBM and normal muscle tissues. |
Immune Infiltration Analysis
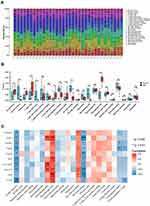
The CIBERSORT algorithm results are shown in Figure 6A. Immune infiltration analysis results of IBM patients and healthy control subjects are shown in Figure 6B. The expression levels of naive B cells, plasma cells, CD8+ T cells, follicular helper T cells, and M2 macrophages were higher in the IBM group than in the healthy control group. Meanwhile, the degree of infiltration of memory B cells, CD4+ naive T cells, and resting natural killer (NK) cells was lower in the IBM group than in the healthy control group. The relationship between the hub genes and the level of the immune cell types was then assessed. STAT1, ITGAM, and CCR5 were positively correlated with M2 macrophages; CXCL10 and CD4 were negatively correlated with the level of resting natural killer (NK) cells (Figure 6C). These results suggested that the hub genes can regulate the immune infiltration of IBM and thus could be potential targets for IBM treatment.
Gene Set Variation and Enrichment Analysis (GSVA and GSEA)
GSVA is used to analyze differences in pathway activities per sample. Herein, GSVA was used to calculate the enrichment scores of Hallmark gene sets in GSE3112 and GSE128470. The enrichment scores of 50 Hallmark gene sets between IBM and normal samples are shown in Figure 7A and B. Multiple pathways and biological processes, including apoptosis, inflammatory response, interferon-alpha response, IL2-STAT5 signaling, and P53 pathways, were activated in IBM.
GSEA analysis was used to explore the potential pathways and biological processes involved in IBM. The GSEA results of GSE3112 showed that genes in the IBM samples were enriched in allograft rejection, apical junction, apoptosis, complement, epithelial-mesenchymal transition, IL2-STAT5 signaling, inflammatory response, interferon-gamma response, KRAS signaling up, and P53 pathway (Figure 7C and D). The GSEA results of GSE128470 showed that genes in the IBM samples were enriched in allograft rejection, inflammatory response, KRAS signaling up, TNFα signaling via NFKβ, complement, epithelial-mesenchymal transition, P53 pathway, interferon-gamma response, apical junction, and IL2-STAT5 signaling (Figure 7E and F). These results indicate that genes in the IBM samples were all enriched in allograft rejection, apical junction, complement, epithelial-mesenchymal transition, IL2-STAT5 signaling, inflammatory response, interferon-gamma response, KRAS signaling up, and P53 pathway. Moreover, the relationship between the hub genes and potential pathways was also explored. The hub genes were positively correlated with these pathways and biological processes (Figure 8A and B).
The DEGs were analyzed using CMap to identify the small molecule drugs, including nadolol, nifuroxazide, 3-acetamidocoumarin, prestwick-1103, and sulfaquinoxaline, with potential therapeutic effects in IBM (Table 1).
 |
Table 1 The Connectivity Map (CMap) Analysis Results |
Discussion
Inclusion body myositis (IBM) has unique characteristics among the idiopathic inflammatory myopathies. However, there are no effective treatment interventions for IBM. It is widely acknowledged that IBM has a complex pathogenesis and has not been fully clarified. It is unclear whether immune-mediated and degenerative processes are causally linked in IBM. Previously studies have reported that IBM pathogenesis involves clonally-restricted and antigen-driven infiltrating CD8-positive T cells. IBM pathogenesis is also associated with abnormal protein deposition and mitochondrial dysfunction. Herein, bioinformatics and data mining analyses were performed to identify the DEGs from muscle biopsy samples from IBM patients and healthy groups. Enrichment analysis was used to screen biochemical processes and pathways. GSVA and GSEA analysis were conducted to confirm the potential pathways of IBM from enrichment analysis. The relative levels of immune cells in IBM and normal muscle groups were also evaluated. In summary, the molecular and immunological characteristics of IBM were investigated, providing possible molecules and signaling pathways for the understanding and treatment of IBM.
Ten hub genes (CD4, PTPRC, ITGAM, TYROBP, CCR5, IRF8, STAT1, ITGB2, HLA-DRA, and CXCL10), closely related to IBM were identified based on PPI network. CD4 encodes a glycoprotein expressed on the surface of T-lymphocyte populations of different functions.15 CD4 expression is correlated with the specificity of T cells to class II or I major histocompatibility complex (MHC) molecules on target cells. Therefore, MHC molecules can modulate immune cells by targeting CD4. Previous research showed that Cytotoxic CD8+T cells, some macrophages, and CD4+T cells could invade the muscle fibers of IBM. PTPRC (CD45) encodes the protein tyrosine phosphatase (PTP) family, regulating T cell and B cell antigen receptor signaling. Moreover, human PTPRC is associated with immunodeficiency disease and viral susceptibility.16 ITGAM encodes integrin αM chains, which play a key role in various inflammatory responses (in monocytes and neutrophils). Some studies have also demonstrated that ITGAM can act as a complement 3 receptor, involved in inflammatory response.14 TYROBP is a transmembrane receptor widely found in macrophages/monocytes, natural killer cells (NK), and neutrophils. Macrophages have played a key role in IBM pathogenesis in recent years. Previous studies have shown that TYROBP is one of the key drivers of multiple inflammatory pathways.17 Meanwhile, IRF8 stabilizes the transcription mechanism of type I IFN promoters, thus enhancing IFN expression in dendritic cells and monocytes.18 These results suggest that immune response is involved in IBM pathogenesis.
CCR5 modulates T-cell migration, and it is associated with some autoimmune disorders. Studies have shown that CCR5 binds up-regulate pro-inflammatory chemokines in IBM,13 consistent with this study. HLA-DRA encodes the subunit HLA-DR of α MHC class II molecule. It is expressed in activated CD4+ T cells, B cells, macrophages, and dendritic cells. Previous studies have shown that MHC class II positivity has high specificity in inflammatory myopathy. Herein, HLA-DRA expression was significantly increased in IBM patients.19 Therefore, it is necessary to analyze the expression pattern and function of HLA-DRA. CXCL10 belongs to CXC chemokine family. CXCL10 induces chemotaxis, apoptosis, and cell growth by binding to CXCR3 receptors. CXCL10 is widely expressed on the surface of IBM myofibers.20 The inflammatory cells invading and surrounding non-necrotic muscle fibers are strongly associated with CCXL10 in the cytotoxic immune response of IBM. Herein, several hub genes (CCR5, HLA-DRA, and PTPRC) could distinguish IBM from other idiopathic inflammatory myopathy, including DM, NM, and NS. In contrast, all the 10 hub genes could not distinguish IBM and PM. However, these results should be verified using large cohorts containing enough samples.
GO and KEGG analyses showed that the DEGs were mainly enriched in Th1 and Th2 cell differentiation, chemokine activity, chemokine signaling pathway, MHC protein complex, antigen processing and presentation, and cellular response to interferon-gamma. These results indicate that immune infiltration and immune response in the local microenvironment play a key role in IBM. Th1-cells can activate the cytotoxicity of macrophages and CD8 T cells, thus inducing cellular immunity. Th2 cells induce the differentiation of B cells into plasma cells or memory B cells, thus producing antibodies. These findings suggest that Th1-mediated immune response is crucial in idiopathic inflammatory myopathy. Moreover, most T cells in muscle tissues are chemokine receptors associated with Th1 lineage. Studies have reported that IFN-γ, a cellular cytokine strongly associated with Th1, is up-regulated in IBM, leading to significant upregulation of autophagy activity in skeletal muscle and amyloid deposition in muscle fibers.20 We found similar results in our research.
Immune cell infiltrations also play a crucial part in IBM pathogenesis. Greenberg reported a group of KLRG1+ T cells that could aggressively intrude IBM myofiber. Therefore, targeting this cell type is a potential treatment for IBM patients.21 Herein, CD8 T cells, Tregs, and macrophages were crucial in IBM. T cells and macrophages are the primary infiltrated immune cells in IBM, similar to this study. Tregs can inhibit effector response mediated by CD8+ T cells, thus protecting muscle cells.22 Herein, the overall degree of Tregs was significantly down-regulated in the IBM group than in the healthy controls. Moreover, the results showed that the hub genes may be involved in the regulation of multiple immune cells in IBM. These findings suggest that targeting hub genes and immune cells might be an effective therapeutic strategy for IBM patients.
The results of GSEA were similar to the DEG functional analysis results. Besides, GSEA revealed various enrichment functions, including IL2-STAT5 signaling, KRAS signaling, and P53 pathway. These signaling pathways are typical mutated pathways in tumorigenesis. IBM has been considered a separate inflammatory myopathy, independent of the tumor, in the past few decades. Although no study has assessed the relationship between IBM and malignancies, some studies have shown the relationship between dermatomyositis and malignant tumor. A retrospective study exhibited a significantly increased incidence of cancer in patients with dermatomyositis.23 In some reports, patients have been diagnosed with uterine cancer, breast cancer, papillary thyroid carcinoma,24 and renal cell carcinoma25 within five years before or after IBM diagnosis. For instance, Michael et al assessed a 64-year-old man with IBM and concurrent transitional cell carcinoma of the bladder who recovered strength and self-care ability.26 However, some rare sporadic IBM may be due to a paraneoplastic presentation. Herein, interferon-gamma response was also identified. Previous studies have reported that the induction of IFN-gamma significantly influences the proliferative response of HIV-infected peripheral blood mononuclear cells in vitro.27 Moreover, the imbalance between the pro-inflammatory and anti-inflammatory responses mediated by polymorphisms in the IFN-gamma may influence the outcome of chronic HCV infection.28 Some researchers have found that IBM may be related to some virus infections, such as hepatitis C virus and HIV virus. A retrospective study investigated the prevalence of HCV infection in IBM patients, and found a markedly higher infection rate of HCV in IBM than that in the age-matched PM.29 It was reported that HIV virus might be a deciding factor for the progress of IBM because over 10% of infiltrating CD8+ T cells displayed specificity for human leukocyte antigen‐A* 0201‐HIV‐gag. However, these views are majorly theoretical.30 Further basic and clinical researches are needed to determine the exact relationship between IBM and these disorders.
There are some limitations in this study. Our study was mainly based on bioinformatics analyses. The in vivo and in vitro researches should be validated to elaborate the molecular and immune-related mechanisms of IBM.
Conclusions
In this study, potential pathways and hub genes (CD4, PTPRC, ITGAM, TYROBP, CCR5, IRF8, STAT1, ITGB2, HLA-DRA, and CXCL10) correlated with IBM were identified through bioinformatics analysis. Further analyses demonstrated that these hub genes can regulate immune infiltrations and multiple pathways in IBM. Finally, CMap database was used to predict five candidate drugs for IBM patients. Therefore, this research can provide new insights into mechanism and potential therapeutic targets for the treatment of IBM.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Schmidt K, Schmidt J. Inclusion body myositis: advancements in diagnosis, pathomechanisms, and treatment. Curr Opin Rheumatol. 2017;29(6):632–638. doi:10.1097/BOR.0000000000000436
2. Souza PVS, Badia BML, Gonçalves EA, Farias IB, Pinto WBVR, Oliveira ASB. Hereditary inclusion body myopathy: a clinical and genetic review. Rev Neurocienc. 2020;28:1–23. doi:10.34024/rnc.2020.v28.10569
3. Callan A, Capkun G, Vasanthaprasad V, Freitas R, Needham M. A Systematic Review and Meta-Analysis of Prevalence Studies of Sporadic Inclusion Body Myositis. J Neuromuscular Dis. 2017;4(2):127–137. doi:10.3233/JND-160198
4. Naddaf E, Barohn RJ, Dimachkie MM. Inclusion Body Myositis: update on Pathogenesis and Treatment. Neurotherapeutics. 2018;15(4):995–1005. doi:10.1007/s13311-018-0658-8
5. Rothwell S, Lilleker JB, Lamb JA. Genetics in inclusion body myositis. Curr Opin Rheumatol. 2017;29(6):639–644. doi:10.1097/BOR.0000000000000431
6. Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi:10.1186/1752-0509-8-S4-S11
7. Xiu MX, Liu YM, Chen GY, Hu C, Kuang BH. Identifying Hub Genes, Key Pathways and Immune Cell Infiltration Characteristics in Pediatric and Adult Ulcerative Colitis by Integrated Bioinformatic Analysis. Dig Dis Sci. 2021;66(9):3002–3014. doi:10.1007/s10620-020-06611-w
8. Cao Y, Tang W, Tang W. Immune cell infiltration characteristics and related core genes in lupus nephritis: results from bioinformatic analysis. BMC Immunol. 2019;20(1):37. doi:10.1186/s12865-019-0316-x
9. Ren C, Li M, Du W, et al. Comprehensive Bioinformatics Analysis Reveals Hub Genes and Inflammation State of Rheumatoid Arthritis. Biomed Res Int. 2020;2020:6943103. doi:10.1155/2020/6943103
10. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi:10.1038/nmeth.3337
11. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi:10.1186/1471-2105-14-7
12. Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. doi:10.1126/science.1132939
13. Rothwell S, Cooper RG, Lundberg IE, et al. Immune-Array Analysis in Sporadic Inclusion Body Myositis Reveals HLA-DRB1 Amino Acid Heterogeneity Across the Myositis Spectrum. Arthritis rheumatology. 2017;69(5):1090–1099. doi:10.1002/art.40045
14. Zirlik A, Maier C, Gerdes N, et al. CD40 ligand mediates inflammation independently of CD40 by interaction with Mac-1. Circulation. 2007;115(12):1571–1580. doi:10.1161/CIRCULATIONAHA.106.683201
15. Littman DR. The structure of the CD4 and CD8 genes. Annu Rev Immunol. 1987;5:561–584. doi:10.1146/annurev.iy.05.040187.003021
16. Hendriks WJ, Pulido R. Protein tyrosine phosphatase variants in human hereditary disorders and disease susceptibilities. Biochim Biophys Acta. 2013;1832(10):1673–1696. doi:10.1016/j.bbadis.2013.05.022
17. Jesse TL, LaChance R, Iademarco MF, Dean DC. Interferon regulatory factor-2 is a transcriptional activator in muscle where It regulates expression of vascular cell adhesion molecule-1. J Cell Biol. 1998;140(5):1265–1276. doi:10.1083/jcb.140.5.1265
18. Jefferies CA. Regulating IRFs in IFN Driven Disease. Front Immunol. 2019;10:325. doi:10.3389/fimmu.2019.00325
19. Scott AP, Laing NG, Mastaglia F, et al. Recombination mapping of the susceptibility region for sporadic inclusion body myositis within the major histocompatibility complex. J Neuroimmunol. 2011;235(1–2):77–83. doi:10.1016/j.jneuroim.2011.02.011
20. De Paepe B, Creus KK, De Bleecker JL. Role of cytokines and chemokines in idiopathic inflammatory myopathies. Curr Opin Rheumatol. 2009;21(6):610–616. doi:10.1097/BOR.0b013e3283317b31
21. Greenberg SA, Pinkus JL, Kong SW, Baecher-Allan C, Amato AA, Dorfman DM. Highly differentiated cytotoxic T cells in inclusion body myositis. Brain. 2019;142(9):2590–2604. doi:10.1093/brain/awz207
22. Waschbisch A, Schwab N, Ruck T, Stenner MP, Wiendl H. FOXP3+ T regulatory cells in idiopathic inflammatory myopathies. J Neuroimmunol. 2010;225(1–2):137–142. doi:10.1016/j.jneuroim.2010.03.013
23. Dalakas MC. Polymyositis, dermatomyositis and inclusion-body myositis. N Engl J Med. 1991;325(21):1487–1498. doi:10.1056/NEJM199111213252107
24. Dardis C, Antezana A, Tanji K, Maccabee PJ. Inclusion Body Myositis: a Case Presenting with Respiratory Failure and Autopsy Findings Leading to the Hypothesis of a Paraneoplastic Cause. Am J Case Rep. 2017;18:700–706. doi:10.12659/AJCR.903566
25. Ytterberg SR, Roelofs RI, Mahowald ML. Inclusion body myositis and renal cell carcinoma. Report of two cases and review of the literature. Arthritis Rheum. 1993;36(3):416–421. doi:10.1002/art.1780360319
26. Jensen ML, Wieting JM, Andary MT, Fankhauser MJ, Jones MJ. Inclusion body myositis and transitional cell carcinoma of the bladder: significant resolution of symptoms after tumor excision. Arch Phys Med Rehabil. 1997;78(3):327–329. doi:10.1016/S0003-9993(97)90043-5
27. Schroecksnadel K, Winkler C, Werner ER, et al. Interferon-gamma-mediated pathways and in vitro PBMC proliferation in HIV-infected patients. Biol Chem. 2009;390(2):115–123. doi:10.1515/BC.2009.018
28. Bouzgarrou N, Hassen E, Farhat K, et al. Combined analysis of interferon-gamma and interleukin-10 gene polymorphisms and chronic hepatitis C severity. Hum Immunol. 2009;70(4):230–236. doi:10.1016/j.humimm.2009.01.019
29. Uruha A, Noguchi S, Hayashi YK, et al. Hepatitis C virus infection in inclusion body myositis: a case-control study. Neurology. 2016;86(3):211–217. doi:10.1212/WNL.0000000000002291
30. Couture P, Malfatti E, Morau G, et al. Inclusion body myositis and human immunodeficiency virus type 1: a new case report and literature review. Neuromuscular Disorders. 2018;28(4):334–338. doi:10.1016/j.nmd.2018.01.005
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.