Back to Journals » Cancer Management and Research » Volume 10
Identification of genes and analysis of prognostic values in nonsmoking females with non-small cell lung carcinoma by bioinformatics analyses
Authors Yang GD, Chen QY, Xiao JM, Zhang HL, Wang ZC, Lin XG
Received 17 May 2018
Accepted for publication 24 August 2018
Published 8 October 2018 Volume 2018:10 Pages 4287—4295
DOI https://doi.org/10.2147/CMAR.S174409
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Harikrishna Nakshatri
Guangda Yang,1 Qianya Chen,1 Jieming Xiao,2 Hailiang Zhang,1 Zhichao Wang,1 Xiangan Lin3
1Department of Cancer Chemotherapy, Zengcheng District People’s Hospital of Guangzhou (BoJi-Affiliated Hospital of Sun Yat-Sen University), Guangzhou, China; 2Department of Emergency, Zengcheng District People’s Hospital of Guangzhou (BoJi-Affiliated Hospital of Sun Yat-Sen University), Guangzhou, China; 3Department of Cancer Chemotherapy, Sun Yat-sen Memorial Hospital of Sun Yat-sen University, Guangzhou, China
Background: This study was performed to identify disease-related genes and analyze prognostic values in nonsmoking females with non-small cell lung carcinoma (NSCLC).
Materials and methods: Gene expression profile GSE19804 was downloaded from the Gene Expression Omnibus (GEO) database and analyzed by using GEO2R. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes were used for the functional and pathway enrichment analysis. Then, the Search Tool for the Retrieval of Interacting Genes, Cytoscape, and Molecular Complex Detection were used to construct the protein–protein interaction (PPI) network and identify hub genes. Finally, the Kaplan–Meier plotter online tool was used for the overall survival analysis of hub genes.
Results: A cohort of 699 differentially expressed genes was screened, and they were mainly enriched in the terms of ECM–receptor interaction, focal adhesion, and cell adhesion molecules. A PPI network was constructed, and 15 hub genes were identified base on the subset of PPI network. Then, two significant modules were detected and several genes were found to be associated with the cell cycle pathway. Finally, nine hub genes’ (UBE2C, DLGAP5, TPX2, CCNB2, BIRC5, KIF20A, TOP2A, GNG11, and ANXA1) expressions were found to be associated with the prognosis of the patients.
Conclusion: Overall, we propose that the cell cycle pathway may play an important role in nonsmoking females with NSCLC and the nine hub genes may be further explored as potential targets for NSCLC diagnosis and treatment.
Keywords: non-small cell lung carcinoma, NSCLC, nonsmoking females, GEO2R, prognostic biomarkers, Kaplan–Meier plotter
Introduction
Lung cancer is one of the leading causes of cancer mortality worldwide.1 Non-small-cell lung cancer (NSCLC), the main type of lung cancer, accounts for 80%–85% of all cases, and small cell lung cancer (SCLC) accounts for around 20% of all cases. Although smoking is the major risk factor for lung cancer,2 only 7% of female patients with lung cancer have a history of cigarette smoking in Taiwan.3 In addition, other factors, such as environmental exposure4,5 or hereditary factors,6 have been reported in association with nonsmoking lung cancer patients. However, the molecular mechanisms of NSCLC among nonsmoking women are unclear, although some genes, such as EML4-ALK,7 EGFR,8 TP53,9 and PIK3CA,10 have been found to be associated with lung cancer in never smokers. Thus, it is important to explore the molecular mechanisms involved in NSCLC’s onset and progression among nonsmoking females and identify effective biomarkers.
Recently, gene microarray and bioinformatics analysis were widely used to identify potential biomarkers of cancer. Interestingly, studies were performed to identify disease-related genes, which were associated with prognosis of breast cancer, by using integrated bioinformatics methods.11,12 Similarly, some studies were performed to find important key genes in lung cancer.13–15 But there are limited studies on nonsmoking females with NSCLC.
In 2010, Lu et al3 obtained a panel of differentially expressed genes (DEGs) from nonsmoking female NSCLC patients and normal samples. Using the same data, the present study was performed to further screen the DEGs and predict their underlying function by functional and pathway enrichment analyses. Then, protein–protein interaction network (PPI) networks and modules of PPI network were performed to identify hub genes. More importantly, the prognostic values of the hub genes were further confirmed by survival analysis in nonsmoking females with NSCLC.
Materials and methods
Microarray data
Microarray expression profiles of GSE19804 were obtained from the Gene Expression Omnibus (GEO) database. GSE19804, which was based on the platform of the GPL570 (Affymetrix Human Genome U133 Plus 2.0 Array, Santa Clara, CA, USA), consisted of 60 samples from nonsmoking female NSCLC patients and 60 normal samples.3
Identification of DEGs
The DEGs between NSCLC samples and normal controls were identified using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/), which is a web tool that is applied to screen DEGs by comparing two groups of samples. |log FC| >1.5 and P<0.01 were selected as the cutoff criterion.
Functional and pathway enrichment analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of DEGs were performed using Database for Annotation, Visualization, and Integrated Discovery (DAVID),16 which is a comprehensive set of functional annotation tools. P<0.05 was set as the cutoff criterion.
PPI network analysis of DEGs and modules selection
The Search Tool for the Retrieval of Interacting Genes (STRING) database17 was used to construct a PPI network for DEGs. Then, the modules of PPI network with significant gene pairs (combined score >0.8) was obtained by the Molecular Complex Detection (MCODE) plugin18 in Cytoscape.19 MCODE scores >14 and the number of nodes >15 were used as the cutoff values. Moreover, the function and pathway enrichment analysis of genes in each module was performed using DAVID.
Survival analysis of the hub genes
Kaplan–Meier plotter is a web tool that predicts the prognostic values of genes in some cancer patients, including breast, ovarian, lung, and gastric cancer patients (http://kmplot.com/analysis/index.php?p=background). The nonsmoking female patients with NSCLC were divided into two groups according to the particular gene expression level (high vs low expression).20 Based on these categories, overall survival (OS) analysis of the two patient groups was then compared by the tool. The HR with 95% CIs and log-rank P-value were calculated and showed.
Results
Identification of DEGs
The total number of samples included 60 NSCLC samples and 60 normal samples. A total of 699 genes were identified, including 161 upregulated and 538 downregulated genes.
GO and KEGG pathway enrichment analysis
We used the DAVID for GO and KEGG pathway enrichment analysis based upon DEGs. The top five GO terms of DEGs are shown in Table 1. As to biological process, the DEGs were significantly enriched in the regulation of cell proliferation, cell adhesion, biological adhesion, vasculature development, and blood vessel development. About cellular component, the DEGs were significantly enriched in the extracellular region part, extracellular region, extracellular space, cell surface, and proteinaceous extracellular matrix. In addition, the most enriched GO terms in molecular function were growth factor binding, carbohydrate binding, calcium ion binding, pattern binding, and polysaccharide binding. On the other hand, the most enriched KEGG pathway terms were as follows: ECM–receptor interaction, focal adhesion, cell adhesion molecules, neuroactive ligand-receptor interaction, and complement and coagulation cascades.
Construction of PPI network and module identification
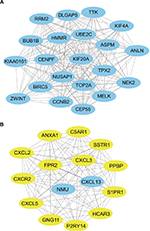
To predict the interactions of the identified DEGs from the protein level, a PPI network from STRING was constructed. In total, 439 nodes and 1,056 edges were shown in the PPI network (Figure 1). The subset of PPI network for the DEGs with a combined score >0.8 was performed to determine the hub genes. The top 15 genes were selected as hub genes, which were GNG11, UBE2C, CCNB2, NMU, ANXA1, FPR2, KIF20A, MELK, NUSAP1, DLGAP5, CENPF, BIRC5, TOP2A, TPX2, and HMMR (Table 2). Subsequently, two modules with MCODE scores >14 and the number of nodes >15 were selected (Figure 2), and enrichment analysis showed that the genes in the modules were mainly associated with cell cycle, chemokine signaling pathway, cytokine-cytokine receptor interaction, and neuroactive ligand-receptor interaction (Table 3).
  | Figure 1 PPI network of DEGs. Abbreviatins: DEGs, differentially expressed genes; PPI, protein–protein interaction. |
  | Table 2 The hub genes that had a degree >20 in PPI network Abbreviation: PPI, protein–protein interaction. |
Survival analysis
Kaplan–Meier plotter was used to predict the prognostic value of 15 identified hub genes. Our results showed that high expression of UBE2C was associated with worse OS for NSCLC patients, as well as DLGAP5, TPX2, CCNB2, BIRC5, KIF20A, and TOP2A (P<0.05) (Figure 3). Additionally, low expression of ANXA1 was associated with poorer OS for NSCLC patients, as well as GNG11 (P<0.05) (Figure 3).
Discussion
In this study, a total of 699 DEGs were screened, including 161 upregulated genes and 538 downregulated genes. Moreover, we selected two significant modules with several key DEGs in the regulatory network of nonsmoking female patients with NSCLC. Then, survival analysis of these genes determined that seven overexpressed genes (UBE2C DLGAP5, TPX2, CCNB2, BIRC5, KIF20A, and TOP2A) and two downregulated genes (GNG11 and ANXA1) were significantly correlated with worse OS of nonsmoking female patients with NSCLC.
The data showed that the seven overexpressed genes are involved in “module 1” of the subnetwork, which is enriched in the cell cycle pathway. As we know that, ubiquitin-conjugating enzyme E2C (UBE2C) is a key regulator of cell cycle progression. High expression of UBE2C is associated with aggressive progression and poor outcome of malignant glioma,21 and UBE2C has been identified as a prognostic protein marker in bladder cancer.22 Moreover, UBE2C, DLGAP5, and TPX2 are associated with the progression and prognosis of pancreatic carcinoma.23 Additionally, both DLGAP5 and TPX2 are mitosis-associated genes and correlated with poor prognosis in NSCLC patients.24 TPX2 overexpression is associated with poor survival in gastric cancer.25 A study also found that TPX2 promotes glioma cell proliferation and invasion via activation of the AKT signaling pathway in glioblastoma multiforme.26 Elevated DLGAP5 expression was negatively correlated with both OS and relapse-free survival of lung cancer.27 A study found that silencing of DLGAP5 by siRNA significantly inhibits the proliferation and invasion of hepatocellular carcinoma cells.28 Cyclin B2 (CCNB2) is a member of cyclin family proteins. Using the same data (GEO:GSE19804), Qian et al further demonstrated that both mRNA and protein expressions of CCNB2 were higher in NSCLC than in normal lung tissues and CCNB2 overexpression is a poor prognostic biomarker in Chinese NSCLC patients.29 BIRC5, which codes for survivin, can participate in cellular survival functions, such as cell cycle progression.30 Moreover, BIRC5 (survivin) is a pejorative prognostic marker in stage II/III breast cancer.31 Similarly, survivin is upregulated in lung cancer tissues and high expression of BIRC5 is associated with poor survival in lung cancer.32,33 Kinesin family member 20A (KIF20A), also known as RAB6KIFL, plays an important role in cytokinesis.34 In addition, it has been reported that high expression of KIF20A is associated with poor OS in cervical squamous cell carcinoma.35 Similarly, a study has found that positive expression of KIF20A indicates poor prognosis of glioma patients.36 High expression of TOP2A, which is involved in DNA synthesis and transcription, is associated with recurrence and metastasis of prostate cancer.37 In addition, high mRNA levels of TOP2A are independent predictors of poor outcome in renal cell carcinoma patients.38 Moreover, another study has found that TOP2A is associated with worse prognosis in NSCLC patients.39 Taken together, these data suggest that UBE2C, DLGAP5, TPX2, CCNB2, BIRC5, KIF20A, and TOP2A are involved in the cell cycle pathway and play a significant role in cancer development, which supports our findings.
On the other hand, GNG11 and ANXA1, which are found in “module 2”, were associated with chemokine signaling pathway, cytokine-cytokine receptor interaction, and neuroactive ligand-receptor interaction. GNG11, a lipid-anchored protein, acts as a tumor suppressor in lung adenocarcinoma.40 Annexin A1 (ANXA1) is a calcium- and phospholipid-binding protein. Some studies found that low ANXA1 expression was associated with better OS in gastric cancer41 and esophageal squamous cell carcinoma.42 However, low expression of ANXA1 was correlated with worse OS in breast cancer patients.43 Downregulation of ANXA1 is correlated with radioresistance in nasopharyngeal carcinoma.44 Moreover, decreased expression of ANXA1 was correlated with poor survival of pancreatic ductal adenocarcinoma (PDAC) patients, and ANXA1 knockdown inhibited cell proliferation, induced G1 phase cell cycle arrest, and increased PDAC cell migration and invasion capacity compared with controls.45 Similarly, ANXA1 knockdown suppressed the proliferation, migration, and invasion of NSCLC cells.46 Together, we speculate that GNG11 and ANXA1 may play a crucial role in NSCLC. ANXA1 could regulate the gastric cancer cell invasion through the formyl peptide receptor (FPR)/extracellular signal-regulated kinase/integrin beta-1-binding protein pathway, and all three FPRs (FPR1 through FPR3) were involved in the regulation process.41 FPR2 promotes invasion and metastasis of gastric cancer cells and predicts the prognosis of patients.47 A study also found that FPR2 was overexpressed in epithelial ovarian cancer (EOC) tissues and FPR2 overexpression indicated poor prognosis of EOC patients.48 However, in the present study, we found that FPR2 was downregulated and low expression indicated better prognosis of NSCLC patients. Similarly, we found that Neuromedin U (Nmu), a secreted neuropeptide, was upregulated and high expression indicated better prognosis of NSCLC patients. But in HNSCC, the expression levels of Nmu in primary tumors with regional metastasis were higher, compared with those without metastasis, and overexpression of Nmu may be involved in the process of regional metastasis of HNSCC.49 So, the roles of FPR2 and Nmu need to be further investigated.
In summary, the present study was intended to identify the potential biomarkers and analyze the prognostic values in nonsmoking females with NSCLC by bioinformatics analysis. We found that hub genes of complex networks, such as UBE2C, DLGAP5, TPX2, CCNB2, BIRC5, KIF20A, TOP2A, GNG11, and ANXA1, may act as potential biomarkers for nonsmoking females with NSCLC. However, the current study was performed by bioinformatics analysis and the conclusions remain to be confirmed by corresponding experiments. Therefore, further investigation is required to verify our findings and determine the potential clinical value of these as biomarkers.
Disclosure
The authors report no conflicts of interest in this work.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. | ||
Wood ME, Kelly K, Mullineaux LG, Bunn PA. The inherited nature of lung cancer: a pilot study. Lung Cancer. 2000;30(2):135–144. | ||
Lu TP, Tsai MH, Lee JM, et al. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers Prev. 2010;19(10):2590–2597. | ||
Lam WK. Lung cancer in Asian women-the environment and genes. Respirology. 2005;10(4):408–417. | ||
Hosgood HD, Boffetta P, Greenland S, et al. In-home coal and wood use and lung cancer risk: a pooled analysis of the International Lung Cancer Consortium. Environ Health Perspect. 2010;118(12):1743–1747. | ||
Wu PF, Lee CH, Wang MJ, et al. Cancer aggregation and complex segregation analysis of families with female non-smoking lung cancer probands in Taiwan. Eur J Cancer. 2004;40(2):260–266. | ||
Wong DW, Leung EL, So KK, et al; University of Hong Kong Lung Cancer Study Group. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR andKRAS. Cancer. 2009;115(8):1723–1733. | ||
Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97(5):339–346. | ||
Toyooka S, Tsuda T, Gazdar AF. The TP53 gene, tobacco exposure, and lung cancer. Hum Mutat. 2003;21(3):229–239. | ||
Yamamoto H, Shigematsu H, Nomura M, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 2008;68(17):6913–6921. | ||
Fang E, Zhang X. Identification of breast cancer hub genes and analysis of prognostic values using integrated bioinformatics analysis. Cancer Biomarkers. 2018;21(2):373–381. | ||
Huang Z, Duan H, Li H. Identification of Gene Expression Pattern Related to Breast Cancer Survival Using Integrated TCGA Datasets and Genomic Tools. Biomed Res Int. 2015;2015:1–10. | ||
Li B-Q, You J, Chen L. Identification of Lung-Cancer-Related Genes with the Shortest Path Approach in a Protein-Protein Interaction Network. Biomed Res Int. 2013;2013(6):1–8. | ||
Jin X, Liu X, Li X, Guan Y. Integrated Analysis of DNA Methylation and mRNA Expression Profiles Data to Identify Key Genes in Lung Adenocarcinoma. Biomed Res Int. 2016;2016:1–9. | ||
Piao J, Sun J, Yang Y, Jin T, Chen L, Lin Z. Target gene screening and evaluation of prognostic values in non-small cell lung cancers by bioinformatics analysis. Gene. 2018;647:306–311. | ||
Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. | ||
Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. | ||
Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. | ||
Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. | ||
Gyo˝rffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. | ||
Ma R, Kang X, Zhang G, Fang F, Du Y, Lv H. High expression of UBE2C is associated with the aggressive progression and poor outcome of malignant glioma. Oncol Lett. 2016;11(3):2300–2304. | ||
Fristrup N, Birkenkamp-Demtröder K, Reinert T, et al. Multicenter validation of cyclin D1, MCM7, TRIM29, and UBE2C as prognostic protein markers in non-muscle-invasive bladder cancer. Am J Pathol. 2013;182(2):339–349. | ||
Zhou Z, Cheng Y, Jiang Y, et al. Ten hub genes associated with progression and prognosis of pancreatic carcinoma identified by co-expression analysis. Int J Biol Sci. 2018;14(2):124–136. | ||
Schneider MA, Christopoulos P, Muley T, et al. AURKA, DLGAP5, TPX2, KIF11 and CKAP5: Five specific mitosis-associated genes correlate with poor prognosis for non-small cell lung cancer patients. Int J Oncol. 2017;50(2):365–372. | ||
Tomii C, Inokuchi M, Takagi Y, et al. TPX2 expression is associated with poor survival in gastric cancer. World J Surg Oncol. 2017;15(1):14. | ||
Gu JJ, Zhang JH, Chen HJ, Wang SS. TPX2 promotes glioma cell proliferation and invasion via activation of the AKT signaling pathway. Oncol Lett. 2016;12(6):5015–5022. | ||
Shi YX, Yin JY, Shen Y, Zhang W, Zhou HH, Liu ZQ. Genome-scale analysis identifies NEK2, DLGAP5 and ECT2 as promising diagnostic and prognostic biomarkers in human lung cancer. Sci Rep. 2017;7(1):8072. | ||
Liao W, Liu W, Yuan Q, et al. Silencing of DLGAP5 by siRNA significantly inhibits the proliferation and invasion of hepatocellular carcinoma cells. PLoS One. 2013;8(12):e80789. | ||
Qian X, Song X, He Y, et al. CCNB2 overexpression is a poor prognostic biomarker in Chinese NSCLC patients. Biomed Pharmacother. 2015;74:222–227. | ||
Blanc-Brude OP, Mesri M, Wall NR, Plescia J, Dohi T, Altieri DC. Therapeutic targeting of the survivin pathway in cancer: initiation of mitochondrial apoptosis and suppression of tumor-associated angiogenesis. Clin Cancer Res. 2003;9(7):2683–2692. | ||
Hamy AS, Bieche I, Lehmann-Che J, et al. BIRC5 (survivin): a pejorative prognostic marker in stage II/III breast cancer with no response to neoadjuvant chemotherapy. Breast Cancer Res Treat. 2016;159(3):499–511. | ||
Yu X, Zhang Y, Cavazos D, et al. miR-195 targets cyclin D3 and survivin to modulate the tumorigenesis of non-small cell lung cancer. Cell Death Dis. 2018;9(2):193. | ||
Duan L, Hu X, Jin Y, Liu R, You Q. Survivin protein expression is involved in the progression of non-small cell lung cancer in Asians: a meta-analysis. BMC Cancer. 2016;16(1):276. | ||
Fontijn RD, Goud B, Echard A, et al. The human kinesin-like protein RB6K is under tight cell cycle control and is essential for cytokinesis. Mol Cell Biol. 2001;21(8):2944–2955. | ||
Zhang W, He W, Shi Y, et al. High Expression of KIF20A Is Associated with Poor Overall Survival and Tumor Progression in Early-Stage Cervical Squamous Cell Carcinoma. PLoS One. 2016;11(12):e167449. | ||
Duan J, Huang W, Shi H. Positive expression of KIF20A indicates poor prognosis of glioma patients. Onco Targets Ther. 2016;9:6741–6749. | ||
Li X, Liu Y, Chen W, et al. TOP2Ahigh is the phenotype of recurrence and metastasis whereas TOP2Aneg cells represent cancer stem cells in prostate cancer. Oncotarget. 2014;5(19):9498. | ||
Chen D, Maruschke M, Hakenberg O, Zimmermann W, Stief CG, Buchner A. TOP2A, HELLS, ATAD2, and TET3 Are Novel Prognostic Markers in Renal Cell Carcinoma. Urology. 2017;102:265.e1–265.e7. | ||
Hou GX, Liu P, Yang J, Wen S. Mining expression and prognosis of topoisomerase isoforms in non-small-cell lung cancer by using Oncomine and Kaplan-Meier plotter. PLoS One. 2017;12(3):e174515. | ||
Hsu YL, Hung JY, Lee YL, et al. Identification of novel gene expression signature in lung adenocarcinoma by using next-generation sequencing data and bioinformatics analysis. Oncotarget. 2017;8(62):104831–104854. | ||
Cheng TY, Wu MS, Lin JT, et al. Annexin A1 is associated with gastric cancer survival and promotes gastric cancer cell invasiveness through the formyl peptide receptor/extracellular signal-regulated kinase/integrin beta-1-binding protein 1 pathway. Cancer. 2012;118(23):5757–5767. | ||
Han G, Tian Y, Duan B, Sheng H, Gao H, Huang J. Association of nuclear annexin A1 with prognosis of patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7(2):751–759. | ||
Wang LP, Bi J, Yao C, et al. Annexin A1 expression and its prognostic significance in human breast cancer. Neoplasma. 2010;57(3):253–259. | ||
Huang L, Liao L, Wan Y, et al. Downregulation of Annexin A1 is correlated with radioresistance in nasopharyngeal carcinoma. Oncol Lett. 2016;12(6):5229–5234. | ||
Liu QH, Shi ML, Bai J, Zheng JN. Identification of ANXA1 as a lymphatic metastasis and poor prognostic factor in pancreatic ductal adenocarcinoma. Asian Pac J Cancer Prev. 2015;16(7):2719–2724. | ||
Fang Y, Guan X, Cai T, et al. Knockdown of ANXA1 suppresses the biological behavior of human NSCLC cells in vitro. Mol Med Rep. 2016;13(5):3858–3866. | ||
Hou XL, Ji CD, Tang J, et al. FPR2 promotes invasion and metastasis of gastric cancer cells and predicts the prognosis of patients. Sci Rep. 2017;7(1):3153. | ||
Xie X, Yang M, Ding Y, Yu L, Chen J. Formyl peptide receptor 2 expression predicts poor prognosis and promotes invasion and metastasis in epithelial ovarian cancer. Oncol Rep. 2017;38(6):3297–3308. | ||
Wang L, Chen C, Li F, et al. Overexpression of neuromedin U is correlated with regional metastasis of head and neck squamous cell carcinoma. Mol Med Rep. 2016;14(2):1075–1082. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.