Back to Journals » Infection and Drug Resistance » Volume 13
Identification of fosA10, a Novel Plasmid-Mediated Fosfomycin Resistance Gene of Klebsiella pneumoniae Origin, in Escherichia coli
Authors Huang Y, Lin Q, Zhou Q, Lv L, Wan M, Gao X, Wang C, Liu JH
Received 26 February 2020
Accepted for publication 7 April 2020
Published 1 May 2020 Volume 2020:13 Pages 1273—1279
DOI https://doi.org/10.2147/IDR.S251360
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Suresh Antony
Ying Huang,1 Qingqing Lin,1 Qiaoli Zhou,1 Luchao Lv,1 Miao Wan,1 Xun Gao,1 Chengzhen Wang,1 Jian-Hua Liu1,2
1Key Laboratory of Zoonoses, Ministry of Agriculture and Rural Affairs, Guangdong Provincial Key Laboratory of Veterinary Pharmaceutics Development and Safety Evaluation, College of Veterinary Medicine, South China Agricultural University, Guangzhou 510642, People’s Republic of China; 2Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou, People’s Republic of China
Correspondence: Jian-Hua Liu Email [email protected]
Purpose: Several subtypes of plasmid-mediated fosfomycin resistance gene fosA in Enterobacteriaceae have been reported worldwide and have caused concern. The present study characterized a novel member of fosA gene located on a plasmid from Escherichia coli.
Materials and Methods: A fosfomycin-resistant E. coli isolate PK9 was recovered from a chicken meat sample in 2018. The presence of fosA genes was detected by PCR and sequencing. Whole-genome sequencing (WGS), conjugation, and cloning were performed to identify the mechanism responsible for fosfomycin resistance. Oxford Nanopore MinION sequencing was carried out to characterize the plasmid carrying fosfomycin resistance gene and the genetic context of the novel fosA variant.
Results: A novel fosA gene with significant homology (> 98%) with fosA6 and fosA5 genes was identified by WGS and was named fosA10. FosA10 shared 56.1% to 98.6% amino acid sequence identity with other reported plasmid-mediated FosA enzymes. Fosfomycin resistance and fosA10 gene were successfully transferred to E. coli C600 by conjugation. Cloning confirmed that FosA10 could confer fosfomycin resistance (MIC > 128 μg/mL). The fosA10 gene was localized on a 53kb IncFII (F35:A-:B-) plasmid. The ∆lysR-fosA10-∆hp fragment (4328 bp), located between two copies of IS10R, showed 100% identity with the chromosomal sequences of 17 Klebsiella pneumoniae strains of ST664 and one of ST3821 in GenBank.
Conclusion: Our findings indicated that the fosA10 gene of E. coli might be captured from the chromosome of K. pneumoniae by IS 10, which further demonstrated that K. pneumoniae might act as a reservoir of fosA-like genes acquired by E. coli.
Keywords: fosfomycin, resistance, plasmid, animal products
Introduction
In recent years, the widespread occurrence of extended-spectrum β-lactamases (ESBLs)-producing and carbapenem-resistant Enterobacteriaceae (CRE) in human clinic has renewed interest in the use of old antimicrobial agents, such as colistin and fosfomycin in the treatment of infections caused by multidrug-resistant pathogens.1–3 Fosfomycin exhibits broad-spectrum bactericidal activity against Gram-positive and Gram-negative bacteria, and is listed as one of the first-line options for the treatment of uncomplicated lower urinary tract infections caused by ESBL-producing Escherichia coli.4,5
Fosfomycin interferes with the biosynthesis of cell wall by inhibiting production of UDP-N-acetylglucosamine enolpyruvyl transferase (MurA).6 Besides, fosfomycin enters the bacterial cell via two transporters, glycerol-3-phosphate transporter (GlpT) and hexose phosphate transporter (UhpT).7,8 Several mechanisms underlying fosfomycin resistance have been identified in E. coli, including modification or overexpression of MurA, inactivation of transporters and their regulatory genes (such as uhpA, cyaA, and ptsI), and acquisition of fosfomycin-modifying enzymes.9 A variety of fosfomycin-modifying enzymes, such as FosA, FosB, FosC, FosL, have been described.10–12 FosA is the most frequently reported group of enzymes in Enterobacteriaceae.12,13 As a glutathione S-transferase, FosA inactivates fosfomycin by catalyzing the addition of glutathione to fosfomycin.13 Until now, nine subtypes of fosA genes have been identified.10,12,14-17 fosA3, the most widespread fosA gene in E. coli, has been detected in human and animal isolates from more than 12 countries, especially in food-producing animal-associated isolates from China.12,18-20 All fosA subtypes, except fosA2 and fosA7, are identified in plasmids, and they are usually originated from the chromosomal gene of Enterobacteriaceae species.14,16,17,21-23
In this study, we reported a novel plasmid-encoded fosA variant, fosA10, in an ESBL-producing E. coli isolate, and characterized the genetic context of fosA10 to identify its origin.
Materials and Methods
Bacterial Isolation
E. coli isolate PK9 was recovered from a local broiler meat outlet in Faisalabad, Pakistan, in March 2018 as previously described.24
Antimicrobial Susceptibility Testing and Detection of fos Genes
The MICs of 13 antimicrobial agents, including ampicillin, cefotaxime, ceftazidime, cefoxitin, florfenicol, streptomycin, doxycycline, ciprofloxacin, imipenem, colistin, amikacin, gentamycin, and tigecycline, were determined by either agar dilution or broth microdilution method (colistin and tigecycline) according to the Clinical Laboratory Standards Institute (CLSI) guideline.25 Besides, the agar dilution method using Mueller-Hinton agar supplemented with 25 μg/mL of glucose-6-phosphate (G6P) was applied to determine the fosfomycin MICs as recommended.25 E. coli ATCC 25922 was used as a quality control standard. The results were interpreted according to the breakpoints of the CLSI.25
The presence of known fos genes (fosA1 to fosA7 and fosC) in PK9 was investigated by PCR amplification as previously described.14,15,22,23,26,27
Whole-Genome Sequencing
Whole genomic DNA of the fosA10-positive E. coli isolate was extracted using HiPure Bacterial DNA Kit (Magen, Guangzhou, China). Whole-genome sequencing (WGS) was performed by Novogene (Beijing Novogene Bioinformatics Co., Ltd., Beijing, China) using Illumina HiSeq 2500 technology (Illumina, San Diego, CA, USA). De novo assembly was performed using SOAPdenovo (version 2.04). To obtain the complete sequence of the fosA10-carrying plasmid, we then sequenced E. coli PK9 on Oxford Nanopore MinION. The assemblies of long Nanopore reads and the short Illumina reads were combined via Unicycler (version 0.4.8). The resulting contigs were uploaded into the Center for Genomic Epidemiology server (https://cge.cbs.dtu.dk/services/). The resistance genes, plasmid type, and multilocus sequence type (MLST) of PK9 were analyzed by ResFinder 3.2, PlasmidFinder, and MLST, respectively.
Conjugation Experiment
Conjugation experiments were performed using isolate PK9 and E. coli strain C600 (high-level resistance to streptomycin) as donor and recipient strains, respectively.28 Transconjugants were selected on MacConkey agar plates containing fosfomycin (64 μg/mL) and streptomycin (3000 μg/mL) for counter-selection.
Cloning of fosA10 Gene
The fosA10 gene from E. coli PK9 was cloned into the pMD19-T vector using primers listed as followed: PK9-F: 5ʹ-TCATTAGGGGATTCATCAGT-3ʹ and PK9-R: 5ʹ-AGATAGTGGAGCGGAGACC-3ʹ. Then, it was transferred into E. coli DH5α (Takara, Shiga, Japan) by heat shock. Transformants were selected on LB agar plates containing 100 μg/mL ampicillin. Recombinant clones were analyzed by PCR followed by Sanger sequencing.
Phylogenic Analyses and Homology Alignment
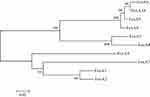
The amino acid sequence of FosA10 was predicted based on the nucleotide sequence of fosA10 gene, and its amino acid sequence was homologously aligned with the amino acid sequence of fosfomycin enzymes in other Enterobacteriaceae obtained from GenBank by T-Coffee.29,30 The phylogenic tree was constructed by distance method using Neighbor-Joining algorithm of MEGA 5. Branch lengths were drawn to scale and were proportional to the number of amino acid substitutions with 500 bootstrap replications.
Results and Discussion
Identification of a Novel Plasmid-Mediated Fosfomycin Resistance Gene fosA10
E. coli PK9 was found to be resistant to ampicillin, cefotaxime, ceftazidime, cefoxitin, streptomycin, gentamycin, and ciprofloxacin, and intermediate to doxycycline (Table 1). Notably, it was highly resistant to fosfomycin with an MIC > 128 μg/mL (Table 1). However, no fosC2 or fosA gene (fosA1–A7) was detected from PK9 by PCR amplification. Conjugation experiment showed that the fosfomycin resistance of PK9 was transferred to the recipient E. coli C600 (Table 1), indicating the presence of plasmid-mediated fosfomycin resistance gene.
 |
Table 1 Antimicrobial Susceptibility of fosA10-Carrying Escherichia coli Strain PK9 and Transconjugant or Transformant |
WGS data showed that E. coli PK9 belonged to ST38. In a 1918 bp contig, a 420 bp open reading frame (ORF) encoding FosA-like protein was identified. This novel fosA gene was then named fosA10 as the next available number according to published data and NCBI.17 fosA10 had 69.7%, 67.8%, 73.4%, 73.1%, 98.3%, 99.3%, 58.2%, 64.0%, 93.9%, and 53.6% nucleotide identity with fosA1 to fosA9 and fosC2, respectively. Besides, the amino acid sequence of FosA10 enzyme encoded by fosA10 shared 70.0%, 68.6%, 79.0%, 78.3%, 98.6%, 97.8%, 62.9%, 65.0%, 97.8%, and 56.1% identity with FosA1 to FosA9 and FosC2, respectively (Figures 1 and S1).
To determine whether fosA10 could confer resistance to fosfomycin, we constructed a recombinant plasmid pMD19-T + fosA10. The MIC of fosfomycin for E. coli DH5α transformed with pMD19-T + fosA10 was >128μg/mL, more than 64-fold higher than that of E. coli DH5α carrying pMD19-T alone (Table 1).
Genetic Context and Origin of the fosA10 Gene
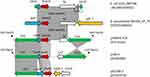
Hybrid assembly of the MinION long reads and HiSeq short reads revealed that PK9 had a circular 5,423,354 bp chromosome and three plasmids (Table 2). One plasmid, designated pHNPK9-FOS, carries fosA10. pHNPK9-FOS was a 53,736 bp plasmid containing 69 predicted ORFs, and belonged to IncFII (F35:A-:B-). It possessed a typical IncF-type backbone, encoding genes for replication, transfer, maintenance, and stability functions (Figure 2). A variable region of 4328 bp consisting of fosA10 and two copies of IS10 element was inserted into a hypothetical gene of the plasmid backbone. The insertion was surrounded by a 9-bp direct repeat sequences (DRs) (TACCTGGTG) suggesting mobilization of this fosA10 gene by composite transposon formed by IS10 (Figure 3). In addition to fosA10 gene, the region surrounded by IS10 includes a truncated transcriptional regulator gene lysR (190 bp) and a 1060 bp sequence (containing a truncated hypothetical gene) located upstream and downstream of fosA10, respectively. BLAST homology analysis demonstrated that the sequence of the 1670 bp region surrounded by IS10 had 100% nucleotide identity with chromosome sequences of 18 K. pneumoniae strains from Austria, USA, UK, Morocco, and Japan (Figure 3 and Table S1), suggesting its mobilization from the chromosome of one K. pneumoniae strain to plasmid. All the 18 K. pneumoniae strains belong to ST644 except one that belongs to ST3821 (Table S1). In addition, a WGS contig (GenBank accession number MUJB01000052) of an E. coli strain AUH_IMP168 collected in 2013 from Lebanon also contained fosA10 and two incomplete IS10 elements caused by short-read DNA sequencing. The fragment containing fosA10 surrounded by IS10 in E. coli strain AUH_IMP168 was also 100% identical to the chromosome sequences of 18 K. pneumoniae strains mentioned above (Figure 3). However, the size of the spacer region between the 3ʹ end of fosA10 and IS10 was longer than that in pHNPK9-FOS (1663 bp vs 1060 bp), indicating occurrence of a different mobilization event of fosA10 from K. pneumoniae chromosome.
 |
Table 2 Resistance Genes and Plasmids Carried by Escherichia coli Strain PK9 |
The other two plasmid-located fosA genes, fosA5 and fosA6, also originated from K. pneumoniae chromosome,14,23 and have high similarity of nucleic acid sequence with fosA10, with difference in only four or seven nucleotides. Like fosA10, lysR genes truncated by IS10 segment were identified upstream of fosA5 (in plasmid pHKU1, GenBank accession number KC960485) and fosA6 (in plasmid pYD786-2, KU254579.1),14,23,31 and the insertion sites of IS10 upstream of fosA5 and fosA10 were the same (Figure 3). In addition, IS10 was found to be inserted downstream of fosA5. However, the fragments located downstream of fosA5, fosA6, and fosA10 were varied. Although fosA5, fosA6, and fosA10 have significant homology and were all derived from the K. pneumoniae chromosome, these mobilization events of fosAkp from the K. pneumoniae chromosome to plasmids seemed to occur separately. Nevertheless, the findings of several events of IS10 insertion in lysR genes of K. pneumoniae in recent years implied that lysR gene was a hotspot for IS10 insertion. Further capture of chromosomal fosA genes from K. pneumoniae by plasmids is possible.
In summary, we report the emergence of a novel plasmid-mediated fosA gene, fosA10, conferring high-level resistance to fosfomycin. fosA10 gene was probably horizontally transferred from K. pneumoniae chromosome to E. coli plasmid by IS10. The increasing discovery of chromosomal fosA genes on plasmids is alarming and demonstrated that Klebsiella spp. might be an important reservoir of fosA genes for E. coli. Considering the apparent horizontal transferability of plasmids, further dissemination of these fosA genes among E. coli is possible and will constitute a serious threat to antimicrobial therapy. Therefore, we should pay more attention to the emergence and spread of plasmid-mediated fosA genes. Further studies on the occurrence and characterization of fosA10-carrying plasmids in Enterobacteriaceae from various sources (humans, animals, and the environment) will offer more insights into the potential role of fosA10 in the dissemination of fosfomycin resistance.
Accession Number
The complete nucleotide sequence of plasmid pHNK9-FOS has been deposited in GenBank under accession no. MT074415.
Acknowledgments
The study was supported in part by grants from the National Natural Science Foundation of China (No. 31625026, 31830099). We would like to thank Zulqarnain Baloch for providing the strain.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Poulakou G, Bassetti M, Righi E, et al. Current and future treatment options for infections caused by multidrug-resistant Gram-negative pathogens. Future Microbiol. 2014;9:1053–1069. doi:10.2217/fmb.14.58
2. Falagas ME, Kastoris AC, Kapaskelis AM, et al. Fosfomycin for the treatment of multidrug-resistant, including extended-spectrum β-lactamase producing, Enterobacteriaceae infections: a systematic review. Lancet Infect Dis. 2010;10:43–50. doi:10.1016/S1473-3099(09)70325-1
3. Sastry S, Doi Y. Fosfomycin: resurgence of an old companion. J Infect Chemother. 2016;22:273–280. doi:10.1016/j.jiac.2016.01.010
4. Patel SS, Balfour JA, Bryson HM. Fosfomycin tromethamine. A review of its antibacterial activity, pharmacokinetic properties and therapeutic efficacy as a single-dose oral treatment for acute uncomplicated lower urinary tract infections. Drugs. 1997;53:637–656. doi:10.2165/00003495-199753040-00007
5. Kanj SS, Kanafani ZA. Current concepts in antimicrobial therapy against resistant gram-negative organisms: extended-spectrum β-lactamase-producing Enterobacteriaceae, carbapenem-resistant enterobacteriaceae, and multidrug resistant Pseudomonas aeruginosa. Mayo Clin Proc. 2011;86:250–259. doi:10.4065/mcp.2010.0674
6. Ito R, Mustapha MM, Tomich AD, et al. Widespread fosfomycin resistance in Gram-negative bacteria attributable to the chromosomal fosA gene. MBio. 2017;8:e00749–17. doi:10.1128/mBio.00749-17
7. Kahan FM, Kahan JS, Cassidy PJ, et al. The mechanism of action of fosfomycin (phosphonomycin). Ann N Y Acad Sci. 1974;235:364–386. doi:10.1111/j.1749-6632.1974.tb43277.x
8. Lemieux MJ, Huang Y, Wang DN. Glycerol-3-phosphate transporter of Escherichia coli: structure, function and regulation. Res Microbiol. 2004;155:623–629. doi:10.1016/j.resmic.2004.05.016
9. Castañeda-García A, Blázquez J, Rodríguez-Rojas A. Molecular mechanisms and clinical impact of acquired and intrinsic fosfomycin resistance. Antibiotics. 2013;2(2):217–236. doi:10.3390/antibiotics2020217
10. Wachino J, Yamane K, Suzuki S, et al. Prevalence of fosfomycin resistance among CTX-M-producing Escherichia coli clinical isolates in Japan and identification of novel plasmid-mediated fosfomycin-modifying enzymes. Antimicrob Agents Chemother. 2010;54:3061–3064. doi:10.1128/AAC.01834-09
11. Kieffer N, Poirel L, Descombes MC, et al. Characterization of FosL1, a plasmid-encoded fosfomycin resistance protein identified in Escherichia coli. Antimicrob Agents Chemother. 2020;64. doi:10.1128/AAC.02042-19
12. Yang T-Y, Lu P-L, Tseng S-P. Update on fosfomycin-modified genes in Enterobacteriaceae. J Microbiol Immunol Infect. 2019;52:9–21. doi:10.1016/j.jmii.2017.10.006
13. Falagas ME, Vouloumanou EK, Samonis GS, et al. Fosfomycin. Clin Microbiol Rev. 2016;29:321–347. doi:10.1128/CMR.00068-15
14. Guo Q, A D T, McElheny CL, et al. Glutathione-S-transferase FosA6 of Klebsiella pneumoniae origin conferring fosfomycin resistance in ESBL-producing Escherichia coli. J Antimicrob Chemother. 2016;71:2460–2465. doi:10.1093/jac/dkw177
15. Rehman MA, Yin X, Persaud-Lachhman MG, et al. First detection of a fosfomycin resistance gene, fosA7, in Salmonella enterica serovar Heidelberg isolated from broiler chickens. Antimicrob Agents Chemother. 2017;61:e00410–e004117. doi:10.1128/AAC.00410-17
16. Poirel L, Vuillemin X, Kieffer N, et al. Identification of FosA8, a Plasmid-encoded fosfomycin resistance determinant from Escherichia coliand its origin in Leclercia adecarboxylata. Antimicrob Agents Chemother. 2019;63:e01403–e01419. doi:10.1128/AAC.01403-19
17. Ten Doesschate T, Abbott IJ, Willems RJL, et al. In vivo acquisition of fosfomycin resistance in Escherichia coli by fosA transmission from commensal flora. J Antimicrob Chemother. 2019;74:3630–3632. doi:10.1093/jac/dkz380
18. Wang J, Zeng ZL, Huang XY, et al. Evolution and comparative genomics of F33: A−: B− plasmids carrying blaCTX-M-55 or blaCTX-M-65 in Escherichia coli and Klebsiella pneumoniae isolated from animals, food products, and humans in China. Msphere. 2018;3:e00137–e001318. doi:10.1128/mSphere.00137-18
19. Yang X, Liu W, Liu Y, et al. F33: A-: B-, IncHI2/ST3, and IncI1/ST71 plasmids drive the dissemination of fosA3 and blaCTX− M− 55/− 14/− 65 in Escherichia coli from chickens in China. Front Microbiol. 2014;5:688. doi:10.3389/fmicb.2014.00688
20. Hou J, Yang X, Zeng Z, et al. Detection of the plasmid encoded fosfomycin resistance gene fosA3 in Escherichia coli of food-animal origin. J Antimicrob Chemother. 2013;68:766–770. doi:10.1093/jac/dks465
21. Tseng SP, Wang SF, Kuo CY, et al. Characterization of fosfomycin resistant extended-spectrum β-lactamase-producing Escherichia coli isolates from human and pig in Taiwan. PLoS One. 2015;10:e0135864. doi:10.1371/journal.pone.0135864
22. Nakamura G, Wachino J, Sato N, et al. Practical agar-based disk potentiation test for detection of fosfomycin-nonsusceptible Escherichia coli clinical isolates producing glutathione-S-transferases. J Clin Microbiol. 2014;52:3175–3179. doi:10.1128/JCM.01094-14
23. Ma Y, Xu X, Guo Q, Wang P, Wang W, Wang M. Characterization of fosA5, a new plasmid-mediated fosfomycin resistance gene in Escherichia coli. Lett Appl Microbiol. 2015;60:259–264. doi:10.1111/lam.12366
24. Baloch Z, Lv L, Yi L, et al. Emergence of almost identical F36: A-: B32 plasmids carrying blaNDM-5 and qepA in Escherichia coli from both Pakistan and Canada. Infect Drug Resist. 2019;12:3981. doi:10.2147/IDR.S236766
25. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; 28th Informational Supplement (M100–S28). Wayne (PA): The Institute; 2018.
26. Hou J, Huang X, Deng Y, et al. Dissemination of the fosfomycin resistance gene fosA3 with CTX-M β-lactamase genes and rmtB carried on IncFII plasmids among Escherichia coli isolates from pets in China. Antimicrob Agents Chemother. 2012;56:2135–2138. doi:10.1128/AAC.05104-11
27. Xu H, Miao V, Kwong W, et al. Identification of a novel fosfomycin resistance gene (fosA2) in Enterobacter cloacae from the Salmon River, Canada. Lett Appl Microbiol. 2011;52:427–429. doi:10.1111/j.1472-765X.2011.03016.x
28. Chen L, Chen ZL, Liu JH, et al. Emergence of RmtB methylase-producing Escherichia coli and Enterobacter cloacae isolates from pigs in China. J Antimicrob Chemother. 2007;59:88–885. doi:10.1093/jac/dkm065
29. Di Tommaso P, Moretti S, Xenarios I, et al. T-coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39:W13–W17. doi:10.1093/nar/gkr245
30. Notredame C, Higgins DG, Heringa J. T-Coffee:A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi:10.1006/jmbi.2000.4042
31. Ho PL, Chan J, Lo WU, et al. Prevalence and molecular epidemiology of plasmid-mediated fosfomycin resistance genes among blood and urinary Escherichia coli isolates. J Med Microbiol. 2013;62:1707–1713. doi:10.1099/jmm.0.062653-0
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.