Back to Journals » International Journal of General Medicine » Volume 14
Identification of circRNA-miRNA-mRNA Regulatory Network and Autophagy Interaction Network in Atrial Fibrillation Based on Bioinformatics Analysis
Authors Chao X , Dai W, Li S, Jiang C, Jiang Z, Zhong G
Received 13 August 2021
Accepted for publication 11 October 2021
Published 19 November 2021 Volume 2021:14 Pages 8527—8540
DOI https://doi.org/10.2147/IJGM.S333752
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Xiaoying Chao,1,* Weiran Dai,1,* Shuo Li,1 Chenyang Jiang,1 Zhiyuan Jiang,2 Guoqiang Zhong1
1Department of Cardiology, The First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi, People’s Republic of China; 2Hypertension Division, The First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Guoqiang Zhong
Department of Cardiology, The First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi, 530021, People’s Republic of China
Tel +86 13978635329
Email [email protected]
Zhiyuan Jiang
Hypertension Division, The First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi, 530021, People’s Republic of China
Tel +86 13597006702
Email [email protected]
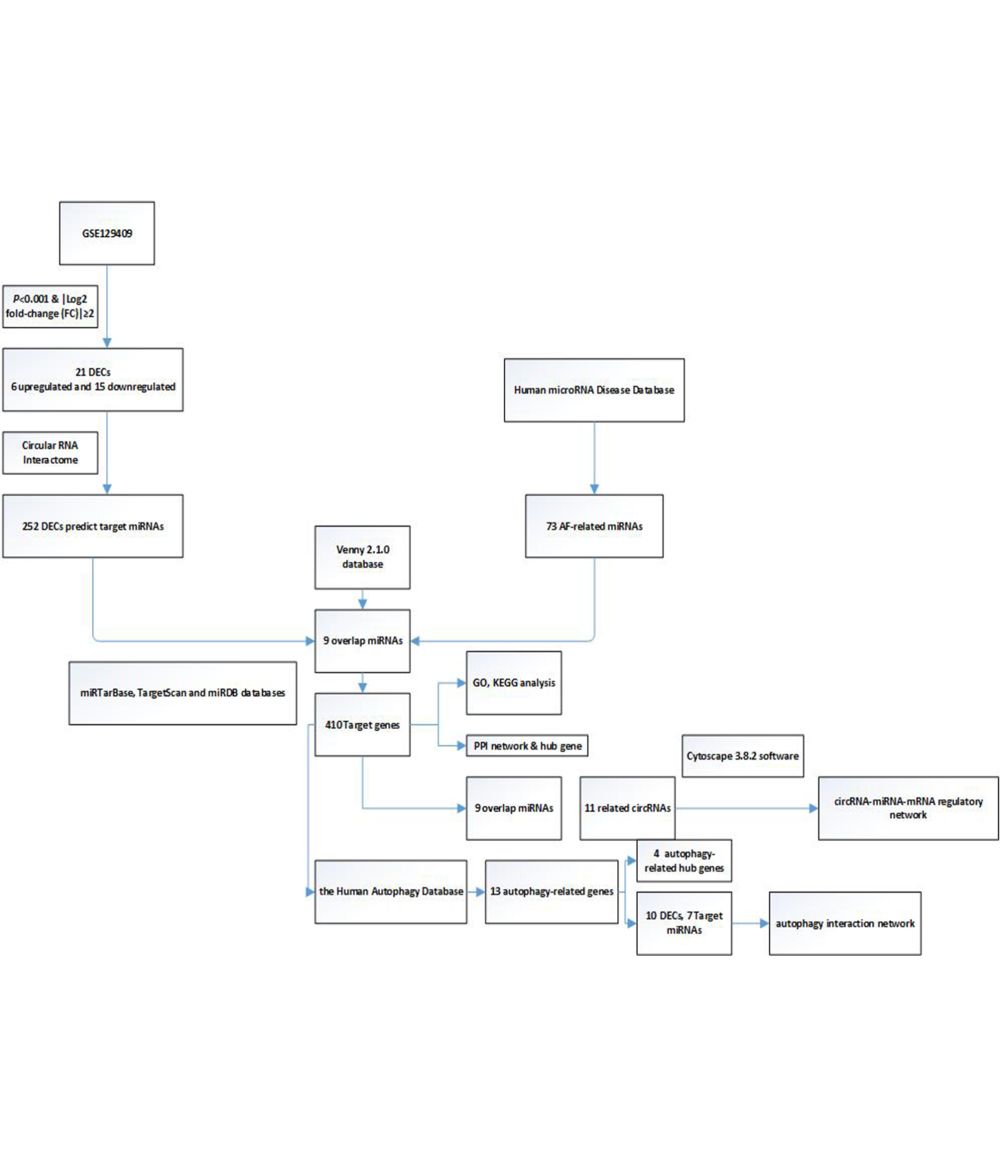
Background: Circular RNA (circRNA) has been receiving increased attention in the research of atrial fibrillation (AF). Our study aims to find potential circRNAs and identify the circRNA-miRNA-mRNA regulatory network in AF based on bioinformatics analysis.
Methods: GSE129409 was retrieved from the Gene Expression Omnibus (GEO) database, and we used R software to analyze the differentially expressed circRNAs (DECs). Subsequently, we used several bioinformatics methods to obtain the target miRNAs and the target genes. Next, we performed Gene Ontology (GO) classification and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the target genes. Then, we used Cytoscape 3.8.2 software to visualize and construct the circRNA-miRNA-mRNA regulatory network, the protein–protein interaction (PPI) network, and the autophagy-related genes network.
Results: We identified a total of 21 DECs, including 6 upregulated DECs and 15 downregulated DECs. After further analysis, we obtained a circRNA-miRNA-mRNA regulatory network consisting of 11 DECs, 9 target miRNAs and 410 target genes, and a PPI network. Finally, the potential novel genes of autophagy in AF were revealed by bioinformatics analysis.
Conclusion: This study could explore the potential role of circRNA, autophagy-related genes and construct the circRNA-miRNA-mRNA regulation network in AF.
Keywords: circular RNA, bioinformatics analysis, ceRNA, autophagy, atrial fibrillation
Graphical Abstract:
Introduction
Atrial fibrillation (AF) is the most common type of arrhythmia in clinical practice, with global prevalence continuing to increase primarily with age.1 In China, 6.5 persons out of every thousand suffer from AF, and there are about 3.9 million patients with AF who are over 60 years old, but it is expected that this number will increase to about 9 million by 2050.2,3 Furthermore, AF may induce or aggravate heart failure, lead to stroke, and have a high risk of disability and fatality. It also poses a burden on the economy and seriously threatened human health and life. In addition to age, factors such as gender, obesity, diet, and lifestyle can affect the prevalence of AF, so mitigating these factors will help to prevent and treat AF.4,5
Non-coding RNA (ncRNA) is not involved in encoding proteins, but it has been discovered to participate in various biological functions such as cell growth, differentiation, apoptosis, and has an indispensable role in cardiovascular diseases.6 Circular RNA (circRNA) is a special non-coding RNA because it lacks the 5ʹ end caps and 3ʹ end poly (A) tails with circular structure.7 Recent research demonstrated that circRNAs could act as competitive endogenous RNAs (ceRNAs) and sponge to bind to miRNAs, regulate transcription, repress or activate gene expression. CeRNAs recently showed to play a crucial role in cardiovascular disease.8 Some latest studies reported that circRNA was related to dilated cardiomyopathy, myocardial infarction, and cardiac hypertrophy.9–11 Similarly, circRNAs were found to take participate in the progression of AF. Multiple circRNAs could relate to valvular AF occurrence and are maintained through miRNAs to construct circRNA-miRNA-mRNA networks.12
Autophagy is a process of self-digestion. In the heart, the basic level of autophagy is vital for maintaining the function and survival of cardiomyocytes, and moderate autophagy has cardioprotective effects.13 Recently, more attention has been paid to the crucial role of autophagy in AF.14 Nevertheless, there are few studies on the autophagy-related regulatory networks in AF.
In this study, we aimed to explore the molecular mechanism of circRNAs and autophagy-related regulatory networks in AF. The current study identified differentially expressed circRNAs (DECs) between patients with AF and healthy controls by analyzing the data in the public Gene Expression Omnibus (GEO) dataset and using the limma package in R software. Subsequently, we used bioinformatics tools to find the target miRNAs and genes. The Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed to detect and verify the functional expression of the target genes. Then, the ceRNA regulatory network (the circRNA-miRNA-mRNA network) and protein–protein interaction (PPI) network were further established. Finally, we focused especially on the target genes associated with autophagy and constructed an autophagy interaction network in which several novel targets were closely associated with AF. Our findings might provide novel information on the role of circRNA and autophagy in AF.
Materials and Methods
Data Source
Gene expression dataset GSE129409 was retrieved for free from Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo).15 The dataset GSE129409 is a circRNA expression array generated using GPL21825 074301 Arraystar Human CircRNA microarray V2. The dataset was collected from human heart tissues from three patients with AF and three healthy controls.
Identification of DECs
The microarray dataset GSE129409 was downloaded, and then the probe name was converted into the international standard circRNA name. The DECs were analyzed by using the limma package in R software (version 4.1.0). The criteria for DECs were adjusted to P value <0.001 and |log fold-change (FC) |≥2.
Prediction of Target miRNAs
Studies showed that there were numerous microRNAs (miRNAs) binding sites in multiple circRNAs, and the circRNAs could affect gene expression via target miRNAs sponge. In this study, we used the Circular RNA Interactome (https://circinteractome.nia.nih.gov/) to predict the target miRNAs of circRNAs.16
AF-Related miRNAs
Human microRNA Disease Database (HMDD) is the comprehensive database of microRNA-disease associations collecting several miRNA-disease association contents.17 We utilized the HMDD database to search for the AF-related miRNAs. Next, we used the Venny 2.1.0 online database (http://bioinfogp.cnb.csic.es/tools/venny/) to analyze the target miRNAs and AF-related miRNAs. If there was an interaction between the analyzed miRNAs mentioned above and the AF-related miRNAs, then they would be closely related to AF. Thus, we could obtain the intersection of these two sets.
Prediction of Target Genes
The intersection of miRNAs were predicted based on three different miRNAs target gene databases, including TargetScan,18 miRTarBase19 and miRDB20 databases. Only these genes were confirmed by all three databases would be considered as the target genes.
Functional Annotation
We used GO annotation (http://www.geneontology.org) and KEGG pathway analysis to determine the functional annotations of the target genes. GO terms with adjust P < 0.05 were selected and integrated using the clusterProfiler package in R software (version 4.1.0). The top 20 enriched GO terms were performed. KEGG pathway analysis was performed to identify the target genes in different biological pathways. The clusterProfiler package in R software (version 4.1.0) was used to conduct the pathways of the target genes. The top 15 enriched KEGG pathways were performed.
Construction of the circRNA-miRNA-mRNA Regulatory Network
The DECs, target miRNAs, and target genes were used to construct a circRNA-miRNA-mRNA regulatory network and visualized by using Cytoscape 3.8.2 software.
PPI Network and Hub Genes Construction
In this study, we used the Search Tool for the Retrieval of Interacting Genes (STRING) database (https://string-db.org/) to construct a PPI network.21 The hub genes were performed by employing plugins of Cytoscape, Molecular Complex Detection (MCODE), and cytoHubba (version 3.8.2).
Construction of an Autophagy Interaction Network
Autophagy genes were identified from the Human Autophagy Database (HADb, http://autophagy.lu/clustering/ index.html). Comparing autophagy genes from the database and the target genes we identified, the intersecting genes were identified as autophagy-related genes. Then, upstream target miRNAs and the autophagy-related genes were performed by the predicted data. The autophagy interaction network was constructed by Cytoscape (version 3.8.2). The hub autophagy-related genes were performed by employing plugins of Cytoscape, Molecular Complex Detection (MCODE), and cytoHubba (version 3.8.2).
Results
DECs Between AF Patients and Healthy Controls
In our study, GSE129409 data was analyzed by the R software, and the criteria for DECs were adjusted to P value <0.001 and |log fold-change (FC) |≥2; 21 DECs were identified. The volcano plots and heatmap for DECs are shown in Figure 1A and B. Among these DECs, six circRNAs were upregulated and 15 circRNAs were downregulated (Table 1).
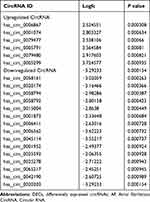 |
Table 1 The DECs Between AF Patients and Healthy Controls |
Identification of Target miRNAs and Target Genes
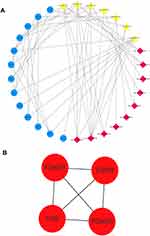
We used the Circular RNA Interactome database to predict the target miRNAs of DECs. The results showed that DECs could regulate 252 target miRNAs. We then obtained 73 AF-related miRNAs from the HMDD database. If the 252 target miRNAs had an interaction with these AF-related miRNAs, the DECs might more likely be related to AF. Therefore, we used the Venn diagram to obtain the intersection between the 252 target miRNAs and AF-related miRNAs, as shown in Figure 2. There were nine overlap miRNAs (hsa-miR-892a, has-miR-451, hsa-miR-377, hsa-miR-223, hsa-miR-215, hsa-miR-192, hsa-miR-183, hsa-miR-155, hsa-miR-144) between these two types of miRNAs, and these overlap miRNAs could be considered as the final target miRNAs. These miRNAs were associated with 11 DECs (Figure 3), and details can be found in Table 2. We predicted the target genes of these final target miRNAs by employing the above 3 databases, and identified 410 target genes.
 |
Table 2 The Target miRNAs and DECs |
 |
Figure 3 The overlap target miRNAs and DECs. The blue nodes represent the overlap target miRNAs and the red and green nodes represent the up- and downregulated DECs, respectively. |
Gene Ontology Analysis and KEGG Analysis
To better comprehend the molecular mechanisms involved in AF, we performed the GO enrichment and KEGG pathway analysis. The enriched GO terms are shown in Figure 4A and B. The top three GO processes included RNA polymerase II-specific, DNA-binding transcription activator activity, and DNA-binding transcription factor binding. In KEGG pathway enrichment analysis, the results showed that KEGG pathway was mainly enriched in signaling pathways regulating pluripotency of stem cells, MAPK signaling pathway, Rap1 signaling pathway, FoxO signaling pathway. The KEGG pathways are shown in Figures 5A and B.
 |
Figure 4 The barplot (A) and dotplot (B) of GO analysis of target genes in ceRNA regulatory network. Abbreviation: GO, Gene Ontology. |
 |
Figure 5 The barplot (A) and dotplot (B) of KEGG analysis of target genes in ceRNA regulatory network. Abbreviation: KEGG, Kyoto Encyclopedia of Genes and Genomes. |
Construction of the circRNA-miRNA-mRNA Regulatory Network
From the previous data analysis, we obtained 11 DECs, 9 overlap target miRNAs, and 410 target genes. Then, we use Cytoscape (Version 3.8.2) to perform the circRNA-miRNA-mRNA relationships in the network (Figure 6).
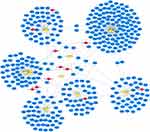 |
Figure 6 The circRNA–miRNA–mRNA regulatory network in AF. The red nodes represent the DECs, the yellow nodes represent the overlap target miRNAs, and the blue nodes represent the target genes. |
Construction of the Protein–Protein Interaction (PPI) Network
We used the identified target genes to construct a PPI network, which consisted of 150 nodes and 291 edges, under the condition that the required interaction score >0.9 and unconnected points were removed (Figure 7A). Nine hub genes (RNF111, ANAPC10, FBXW7, CUL3, BTBD1, TRIM32, CDC27, UBE2H and RNF123) were identified from the PPI network when the degrees ≥8 (Figure 7B).
Construction of the Autophagy Interaction Network
We constructed an autophagy interaction network using autophagy-related genes. A total of 10 DECs, 7 target miRNAs and 13 target genes were involved in the network (Figure 8A). In the autophagy interaction network, four hub genes among the above autophagy-related genes were identified if the degrees ≥4 (Figure 8B).
Discussion
Recently, an increasing number of studies have shown that ncRNA was no longer considered as “transcription junk” as it has an indispensable role in the occurrence and development of AF. In recent years, circRNA is characterized by natural stability, even more, stable than linear RNA in cardiac diseases.22 Researchers found that circRNA was linked to coronary artery disease, myocardial infarction, and AF.23–25 Gao et al determined that hsa_circ_0004104 could be a potential biomarker in AF persistency via targeting MAPK and TGF-β pathways.26 circRNA also has a similar biological function to long non-coding RNA, which can sponge miRNA, thereby reducing the inhibition of miRNA on downstream target genes and upregulating the expression of target genes.27 Therefore, circRNA’s expression and circRNA-miRNA-mRNA network regulation have been increasingly reported in cardiac diseases. Deng et al showed the module of circ-HIPK3-miR-17-3p-ADCY6 axis and circ-HIPK3 might be a therapeutic target of heart failure.28 Huang et al demonstrated that the circ_0001946- miR-7-5p-PARP1 could be a regulatory axis that might contribute to the coronary atherosclerotic heart diseases development.29
In our study, the duration of three AF patients was greater than 1 year, which called persistent AF (pAF). We used bioinformatics and the GEO database to identify differentially expressed circRNAs in patients with AF and healthy controls. The results obtained 21 DECs, including 6 upregulated DECs and 15 downregulated DECs. Among the DECs, Wen et al found that circ_0006867 was a key circRNA in non-small cell lung cancer patients, and it could relate to acquired resistance of tyrosine kinase inhibitor of epidermal growth factor receptor (EGFR).30 Wen and collaborators indicated that circ_0079480 might function as a new treatment target for acute myeloid leukemia therapy.31 Cen et al confirmed that circ_0015004 had a potential oncogenic role in renal cell carcinoma.32 Therefore, the role of these circRNAs in AF had not been reported. Through our research, we speculated that these circRNAs may play a rigorous role in the progression of pAF.
Next, we obtained 9 related-AF target miRNAs, their corresponding 11 circRNAs are more likely to link to AF. Hu and coworkers showed that quercetin could inhibit the expression of miR-223-3p in AF model cells and tissues, and enhance the expression of FOXO3 and activate autophagy, and significantly inhibit myocardial fibrosis, and improve myocardial remodeling in AF.33 Wang et al determined that miR-155 could target the α1c subunit of the calcium channel (CACNA1C) and regulate electrical remodeling in AF.34 Then, a total of 410 potential target genes were associated with the identified target genes, which were predicted using the 3 distinct miRNA target gene prediction databases. In the GO annotation analysis, the top three enriched terms were RNA polymerase II-specific, DNA-binding transcription activator activity, and DNA-binding transcription factor binding. In KEGG pathway enrichment analysis, the signaling pathways regulating pluripotency of stem cells, MAPK signaling pathway, Rap1 signaling pathway, FOXO signaling pathway, and pathways in cancer were mainly enriched.
Among them, the MAPK signaling pathway is a significant energy metabolism pathway related to AF. In a dog model, MAPK activation contributed to arrhythmogenic atrial structural remodeling.35 In atrial myocytes, previous evidence showed that cytoplasmic Ca2+ levels affected the MAPK pathway and caused ion channel remodeling.36 It is considered a cardiac metabolic regulator. Interestingly, Samanta and collaborators showed that oleic acid-activated AMPK and increased nuclear FOXO1 localization, thereby reducing NFATc3 expression and attenuating cardiomyocyte hypertrophy.37 FOXO1 is a key factor of the FOXO signaling pathway, so the MAPK pathway and FOXO signaling pathway may be involved in the occurrence of myocardial remodeling.
We constructed circRNA-miRNA-mRNA and PPI networks. Nine hub genes of the regulatory network, including CDC27, RNF111, CUL3, ANAPC10, FBXW7, UBE2H, BTBD1, RNF123, TRIM32, were identified. miR-223 could bind to FBXW7 in cancers.38 In addition, Yang et al indicated that miR-223 could target FBXW7 in the AKT signaling pathway to regulate physiological cardiac hypertrophy. RNF123 and TRIM32 are ubiquitin E3-ligases.39 Wang and collaborators found that miR-155-5p modulated RNF123 expression in modulating downstream NF-κB1 targets in glioblastoma progression.40 Fatima et al reported that TRIM32 was targeted by miR-155 to modulate human neural stem cell proliferation.41 Therefore, these regulatory networks had not been reported in AF.
Autophagy is a vital cellular process that regulates the clearance of misfolded and proteotoxic proteins, therefore, excessive autophagy may lead to non-apoptotic programmed cell death.42 Impaired autophagy or excessive autophagy can lead to diseases. Nishida and coworkers suggested that impaired autophagosome clearance were involved increased cardiomyocyte death in myocardial ischemia and reperfusion.43 Recently, increasing evidence found that autophagy may be involved in the pathogenesis of AF. The relationship between autophagy and AF is interesting to explore. Exploring the levels of autophagy-related genes may help to study how autophagy affects diseases. However, there are still few related studies in AF, and further investigations will be required to better understand this field. The results of our study contributed to our understanding of genes in the autophagy interaction network, especially those defined as hub genes, including EGFR, FOS, FOXO1, FOXO3. In recent years, a mushrooming number of circRNAs were found to participate in the occurrence and development of diseases through autophagy. Our study found that 10 circRNAs regulated AF in circRNA-miRNA-mRNA autophagy-related networks. These circRNAs might play an important role in the occurrence or maintenance of pAF.
EGFR has a fundamental role in normal physiology and disease, and it is a reasonable target for disease treatment. Hasin et al studies that EGFR is activated in response to angiotensin II and might be involved in left atrial-generated pathology.44 Increasing evidence were reported the involvement of EGFR pathway activation in the development of cardiac hypertrophy.45 Yan et al suggested that Gallic acid could activate autophagy and then induce the degradation of EGFR in cardiac hypertrophic remodeling.46 Enhanced autophagocytosis associated with pathological activation of cardiac fibroblasts. Studies have reported that fos-related antigen 2 regulates autophagocytosis and controls the differentiation of cardiac fibroblasts.47 To our best knowledge, Atrial fibrosis is considered to be the basis of pAF and a sign of structural remodeling. So FOS may be considered to involve in AF. FOXO1, FOXO3 are genes of the FOXO subfamily. As we know, researches in recent years have identified the FOXO subfamily as key players in regulating diverse cellular processes linked to cardiac aging and diseases. In addition, FOXO factors have been shown to play a key role in mediating the effects of factors like transforming growth factor beta-1 (TGF-β1).48 Sengupta et al found that overexpression of either FOXO1 or FOXO3 reduced cardiomyocyte cell size and induced expression of autophagy pathway genes.49 In summary, we inferred that these autophagy-related genes might have significant roles in the occurrence of AF. However, the explicit mechanism of these autophagy-related genes in AF remains largely unclear. We intend to explore more potential autophagy-related genes of AF in the future.
However, there are still some limitations in this study. First, the differential expression of circRNAs we obtained were from only one data set. And target miRNAs and their target genes were a bit weak as the proposed mechanism was based on prediction. In addition, we need to validate our conclusion by future experiments and further research.
Conclusion
In summary, we used different bioinformatics methods to construct a circRNA-miRNA-mRNA network consisting of 11 DECs, 9 target miRNAs, and 410 target genes. Moreover, this study showed that the autophagy interaction network, which we constructed participates extensively in AF. Our study provided new insight into AF at the molecular level from the perspective of the circRNA-miRNA-mRNA regulatory network. This may help the diagnosis and treatment of clinical intervention targets.
Abbreviations
CircRNA, circular RNA; AF, atrial fibrillation; GEO, Gene Expression Omnibus; DECs, differentially expressed circRNAs; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; NcRNA, non-coding RNA; ceRNAs, competitive endogenous RNAs; PPI, protein–protein interaction; miRNAs, microRNAs; HMDD, Human microRNA Disease Database; STRING, Search Tool for Recurring Instances of Neighbouring Genes; MCODE, Molecular Complex Detection; HADb, Human Autophagy Database.
Data Sharing Statement
The GSE129409 dataset is available from the Gene Expression Omnibus GEO (http://www.ncbi.nlm.nih.gov/geo).
Acknowledgments
We would like to acknowledge the reviewers and editors for their invaluable comments on this study.
Funding
This study was funded by the National Natural Science Foundation of China (No. 82060068).
Disclosure
The authors declare that they have no competing interests.
References
1. Kannel W, Abbott R, Savage D, McNamara P. Epidemiologic features of chronic atrial fibrillation: the Framingham study. N Engl J Med. 1982;306(17):1018–1022. doi:10.1056/NEJM198204293061703
2. Zhou Z, Hu D. An epidemiological study on the prevalence of atrial fibrillation in the Chinese population of mainland China. J Epidemiol. 2008;18(5):209–216. doi:10.2188/jea.JE2008021
3. Rahman F, Kwan G, Benjamin E. Global epidemiology of atrial fibrillation. Nat Rev Cardiol. 2014;11(11):639–654. doi:10.1038/nrcardio.2014.118
4. Nalliah C, Sanders P, Kalman J. The impact of diet and lifestyle on atrial fibrillation. Curr Cardiol Rep. 2018;20(12):137. doi:10.1007/s11886-018-1082-8
5. Díez-Villanueva P, Alfonso F. Atrial fibrillation in the elderly. J Geriatric Cardiol. 2019;16(1):49–53.
6. Beermann J, Piccoli M, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–1325. doi:10.1152/physrev.00041.2015
7. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi:10.1261/rna.035667.112
8. Fan X, Weng X, Zhao Y, Chen W, Gan T, Xu D. Circular RNAs in cardiovascular disease: an overview. Biomed Res Int. 2017;2017:1–9. doi:10.1155/2017/5135781
9. Lin Z, Zhao Y, Dai F, Su E, Li F, Yan Y. Analysis of changes in circular RNA expression and construction of ceRNA networks in human dilated cardiomyopathy. J Cell Mol Med. 2021;25(5):2572–2583. doi:10.1111/jcmm.16251
10. Si X, Zheng H, Wei G, et al. circRNA Hipk3 induces cardiac regeneration after myocardial infarction in mice by binding to Notch1 and miR-133a. Mol Ther Nucleic Acids. 2020;21:636–655. doi:10.1016/j.omtn.2020.06.024
11. Li H, Xu J, Fang X, et al. Circular RNA circRNA_000203 aggravates cardiac hypertrophy via suppressing miR-26b-5p and miR-140-3p binding to Gata4. Cardiovasc Res. 2020;116(7):1323–1334. doi:10.1093/cvr/cvz215
12. Zhu X, Tang X, Chong H, et al. Expression profiles of circular RNA in human atrial fibrillation with valvular heart diseases. Front Cardiovasc Med. 2020;2:7.
13. Nakai A, Yamaguchi O, Takeda T, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13(5):619–624. doi:10.1038/nm1574
14. Garcia L, Verdejo H, Kuzmicic J, et al. Impaired cardiac autophagy in patients developing postoperative atrial fibrillation. J Thorac Cardiovasc Surg. 2012;143(2):451–459. doi:10.1016/j.jtcvs.2011.07.056
15. Barrett T, Troup D, Wilhite S, et al. NCBI GEO: mining tens of millions of expression profiles–database and tools update. Nucleic Acids Res. 2007;35:760–765. doi:10.1093/nar/gkl887
16. Dudekula D, Panda A, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13(1):34–42. doi:10.1080/15476286.2015.1128065
17. Huang Z, Shi J, Gao Y, et al. HMDD v3.0: a database for experimentally supported human microRNA-disease associations. Nucleic Acids Res. 2019;47:1013–1017. doi:10.1093/nar/gky1010
18. Agarwal V, Bell G, Nam J, Bartel D. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;1;4.
19. Huang H, Lin Y, Li J, et al. miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020;48:148–154.
20. Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:127–131. doi:10.1093/nar/gkz757
21. Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:447–452. doi:10.1093/nar/gku1003
22. Wang Y, Liu B. Circular RNA in diseased heart. Cells. 2020;9(5):1240. doi:10.3390/cells9051240
23. Wang L, Shen C, Wang Y, et al. Identification of circular RNA Hsa_circ_0001879 and Hsa_circ_0004104 as novel biomarkers for coronary artery disease. Atherosclerosis. 2019;286:88–96. doi:10.1016/j.atherosclerosis.2019.05.006
24. Huang S, Li X, Zheng H, et al. Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. 2019;139(25):2857–2876. doi:10.1161/CIRCULATIONAHA.118.038361
25. Costa M, Kurc S, Drożdż A, Cortez-Dias N, Enguita F. The circulating non-coding RNA landscape for biomarker research: lessons and prospects from cardiovascular diseases. Acta Pharmacol Sin. 2018;39(7):1085–1099. doi:10.1038/aps.2018.35
26. Gao Y, Liu Y, Fu Y, et al. The potential regulatory role of hsa_circ_0004104 in the persistency of atrial fibrillation by promoting cardiac fibrosis via TGF-β pathway. BMC Cardiovasc Disord. 2021;21(1):25. doi:10.1186/s12872-021-01847-4
27. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-7
28. Deng Y, Wang J, Xie G, Zeng X, Li H. Circ-HIPK3 strengthens the effects of adrenaline in heart failure by MiR-17-3p - ADCY6 Axis. Int J Biol Sci. 2019;15(11):2484–2496. doi:10.7150/ijbs.36149
29. Huang S, Zeng Z, Sun Y, et al. Association study of hsa_circ_0001946, hsa-miR-7-5p and PARP1 in coronary atherosclerotic heart disease. Int J Cardiol. 2021;328:1–7. doi:10.1016/j.ijcard.2020.12.026
30. Wen C, Xu G, He S, et al. Screening circular RNAs related to acquired gefitinib resistance in non-small cell lung cancer cell lines. J Cancer. 2020;11(13):3816–3826. doi:10.7150/jca.39783
31. Hu Q, Gu Y, Chen S, Tian Y, Yang S. Hsa_circ_0079480 promotes tumor progression in acute myeloid leukemia via miR-654-3p/HDGF axis. Aging. 2020;13(1):1120–1131. doi:10.18632/aging.202240
32. Cen J, Liang Y, Huang Y, et al. Circular RNA circSDHC serves as a sponge for miR-127-3p to promote the proliferation and metastasis of renal cell carcinoma via the CDKN3/E2F1 axis. Mol Cancer. 2021;20(1):19. doi:10.1186/s12943-021-01314-w
33. Hu J, Wang X, Cui X, Kuang W, Li D, Wang J. Quercetin prevents isoprenaline-induced myocardial fibrosis by promoting autophagy via regulating miR-223-3p/FOXO3. Cell Cycle. 2021;2:1–17.
34. Wang J, Ye Q, Bai S, et al. Inhibiting microRNA-155 attenuates atrial fibrillation by targeting CACNA1C. J Mol Cell Cardiol. 2021;155:58–65. doi:10.1016/j.yjmcc.2021.02.008
35. Li D, Shinagawa K, Pang L, et al. Effects of angiotensin-converting enzyme inhibition on the development of the atrial fibrillation substrate in dogs with ventricular tachypacing-induced congestive heart failure. Circulation. 2001;104(21):2608–2614. doi:10.1161/hc4601.099402
36. Cheng W, Zhu Y, Wang H. The MAPK pathway is involved in the regulation of rapid pacing-induced ionic channel remodeling in rat atrial myocytes. Mol Med Rep. 2016;13(3):2677–2682. doi:10.3892/mmr.2016.4862
37. Samanta J, Mondal A, Saha S, Chakraborty S, Sengupta A. Oleic acid protects from arsenic-induced cardiac hypertrophy via AMPK/FoxO/NFATc3 pathway. Cardiovasc Toxicol. 2020;20(3):261–280. doi:10.1007/s12012-019-09550-9
38. Eto K, Iwatsuki M, Watanabe M, et al. The sensitivity of gastric cancer to trastuzumab is regulated by the miR-223/FBXW7 pathway. Int j Cancer. 2015;136(7):1537–1545. doi:10.1002/ijc.29168
39. Yang L, Li Y, Wang X, et al. Overexpression of miR-223 tips the balance of pro- and anti-hypertrophic signaling cascades toward physiologic cardiac hypertrophy. J Biol Chem. 2016;291(30):15700–15713. doi:10.1074/jbc.M116.715805
40. Wang X, Bustos MA, Zhang X, et al. Downregulation of the ubiquitin-E3 Ligase RNF123 promotes upregulation of the NF-kappaB1 Target SerpinE1 in aggressive glioblastoma tumors. Cancers. 2020;12(5):2548.
41. Fatima M, Kumari R, Schwamborn J, et al. Tripartite containing motif 32 modulates proliferation of human neural precursor cells in HIV-1 neurodegeneration. Cell Death Differ. 2016;23(5):776–786. doi:10.1038/cdd.2015.138
42. Li J, Zhang D, Wiersma M, Brundel B. Role of autophagy in proteostasis: friend and foe in cardiac diseases. Cells. 2018;7:12. doi:10.3390/cells7120279
43. Ma X, Liu H, Foyil S, et al. Impaired autophagosome clearance contributes to cardiomyocyte death in ischemia/reperfusion injury. Circulation. 2012;125(25):3170–3181. doi:10.1161/CIRCULATIONAHA.111.041814
44. Hasin T, Elhanani O, Abassi Z, Hai T, Aronheim A. Angiotensin II signaling up-regulates the immediate early transcription factor ATF3 in the left but not the right atrium. Basic Res Cardiol. 2011;106(2):175–187. doi:10.1007/s00395-010-0145-9
45. Esposito G, Perrino C, Cannavo A, et al. EGFR trans-activation by urotensin II receptor is mediated by β-arrestin recruitment and confers cardioprotection in pressure overload-induced cardiac hypertrophy. Basic Res Cardiol. 2011;106(4):577–589. doi:10.1007/s00395-011-0163-2
46. Yan X, Zhang Y, Zhang L, et al. Gallic acid suppresses cardiac hypertrophic remodeling and heart failure. Mol Nutr Food Res. 2019;63(5):e1800807. doi:10.1002/mnfr.201800807
47. Seidenberg J, Stellato M, Hukara A, et al. The AP-1 Transcription factor Fosl-2 regulates autophagy in cardiac fibroblasts during myocardial fibrogenesis. Int J Mol Sci. 2021;22(4):1861. doi:10.3390/ijms22041861
48. Mishra S, Ravi V, Sundaresan NR. Role of FoxO transcription factors in aging-associated cardiovascular diseases. Vitam Horm. 2021;115:449–475.
49. Sengupta A, Molkentin JD, Yutzey KE. FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem. 2009;284(41):28319–28331. doi:10.1074/jbc.M109.024406
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.