Back to Journals » Cancer Management and Research » Volume 10
Identification of chemoresistance-associated miRNAs in breast cancer
Authors Lou W, Liu J, Ding B, Xu L, Fan W
Received 6 May 2018
Accepted for publication 8 August 2018
Published 23 October 2018 Volume 2018:10 Pages 4747—4757
DOI https://doi.org/10.2147/CMAR.S172722
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Antonella D'Anneo
Weiyang Lou,1–3,* Jingxing Liu,4,* Bisha Ding,1–3 Liang Xu,1–3 Weimin Fan1–3,5
1Program of Innovative Cancer Therapeutics, Division of Hepatobiliary and Pancreatic Surgery, Department of Surgery, First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou 310003, China; 2Key Laboratory of Organ Transplantation, Zhejiang Province, Hangzhou 310003, China; 3Key Laboratory of Combined Multi-organ Transplantation, Ministry of Public Health, Hangzhou 310000, China; 4Department of Intensive Care Unit, Changxing People’s Hospital of Zhejiang, Zhejiang Province, Huzhou 313100, China; 5Department of Pathology and Laboratory Medicine, Medical University of South Carolina, Charleston, SC 29425, USA
*These authors contributed equally to this work
Background: Neoadjuvant chemotherapy (NAC) is an effective therapeutic regimen for patients with breast cancer. However, some individuals cannot benefit from NAC because of drug resistance. To date, valid strategies about enhancing sensitivity of breast cancer to NAC are still scarce. miRNAs have been reported to proverbially be involved in the onset and development of malignancies including drug resistance.
Methods: GSE73736 was downloaded from the GEO database. Student’s t-test was conducted to acquire differentially expressed-miRNAs (DE-miRNAs). Potential target genes of DE-miRNAs were predicted by miRTarBase. Gene Ontology annotation and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses for these target genes were performed by database for annotation, visualization, and integrated discovery. Protein–protein interaction network was constructed by STRING database and visualized through Cytoscape software. The hub target gene-miRNA network was also established by Cytoscape software. Next, the expression of potential functional miRNAs in breast cancer cell lines and tissues was determined. Finally, the roles of miR-3617-3p, miR-3136-3p, and miR-520b in modulating breast cancer chemoresistance were further examined.
Results: A total of 123 DE-miRNAs were identified, including 60 upregulated miRNAs and 63 downregulated miRNAs in the chemoresistant breast cancer group when compared with the chemosensitive group. Six hundred and seventeen and 1,146 potential target genes for the top 10 most upregulated and downregulated miRNAs were predicted, respectively. Enrichment analyses revealed that these target genes were enriched in some cancer-associated or chemoresistance-associated pathways, such as MAPK signaling pathway, wnt signaling pathway, and p53 signaling pathway. MAPK1 and PRDM10 were identified as hub genes in the protein–protein interaction network. The top 25 hub genes were potentially regulated by 16 DE-miRNAs, among which miR-3617-3p and miR-3136-3p were commonly upregulated, whereas miR-520b was downregulated in two chemoresistant breast cancer cells compared with chemosensitive cell. By analyzing TCGA data, we found that expression of miR-3136-3p and miR-520b was increased and decreased in breast cancer tissues, respectively. Moreover, functional experiments demonstrated that miR-3136-3p and miR-3617-3p could reduce chemosensitivity of breast cancer, whereas miR-520b could reverse chemoresistance.
Conclusion: The present study, based on bioinformatics analysis and experimental validation, brings to light novel mechanisms of breast cancer NAC resistance.
Keywords: breast cancer, chemoresistance, neoadjuvant chemotherapy miRNA, bioinformatics analysis, enrichment analysis
Introduction
Breast cancer is one of the most common malignancies among women, and its morbidity is growing yearly in most countries including America. In 2017, according to the statistics provided by the American Cancer Society, more than 250,000 new cases of breast cancer and ~40,000 breast cancer deaths are expected to occur.1 Surgery, radiotherapy, and chemotherapy are still three major traditional treatments for breast cancer. In the past decades, neoadjuvant chemotherapy (NAC) has been widely applied for improving disease free and overall survival and enabling more limited breast-conserving surgery in patients with breast cancer,2 and has substantially enhanced the efficacy of treatment for breast cancer. However, the benefit to patients with breast cancer from NAC is extremely limited in some individuals due to drug resistance.3 It is considerably meaningful to search and develop an approach which can reverse the drug resistance or increase the chemosensitivity of NAC in breast cancer.
MiRNAs are a group of small, nature-occurred, noncoding RNAs with ~18–24 nucleotides in length. MiRNAs regulate gene expression, mainly by binding to the 3′ untranslated region of target mRNA at posttranscriptional level, thereby causing direct degradation of mRNA or inhibition of protein translation.4 Through this approach, miRNAs are involved in a wide range of biological processes (BPs), such as proliferation, differentiation, apoptosis, autophagy, migration, and invasion.5 Many studies have demonstrated that miRNAs play key roles in the occurrence and progression of malignancies including breast cancer.6 Recently, miRNAs have been reported to be associated with chemoresistance of breast cancer.7–9 However, studies regarding the impact of miRNAs in regulating breast cancer NAC resistance are still rare. In this study, we successfully identified several chemoresistance-associated miRNAs (miR-3617-3p, miR-3136-3p, and miR-520b) in breast cancer via a series of processes, including identification of differentially expressed-miRNAs (DE-miRNAs), functional and pathway enrichment analysis, construction and analysis of the protein–protein interaction (PPI) network and miRNA-target gene network, and experimental validation in cell lines. The three miRNAs, miR-3617-3p, miR-3136-3p, and miR-520b, may be important novel NAC response predictors, prognosticators, or therapeutic targets in breast cancer in the future.
Materials and methods
MiRNA microarray
The microarray data set GSE73736 based on GPL20986 MRA-1001B1 Human miRNA array miRbase release 19 was downloaded from the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo). This data set contained 20 cases of breast cancer tissues. According to Mille-Payne response, these 20 cases included 10 cases of NAC-resistant breast cancer tissues (GSM1901882–GSM1901891) which were MP1, and 10 cases of NAC-sensitive breast cancer tissues (GSM1901892–GSM1901901) which were MP5. The MP1 group represents that breast cancer in this group is totally unresponsive to NAC, whereas the MP5 group indicates that breast cancer in this group disappears after administration of NAC.
Screening for DE-miRNAs
Data were first normalized by the normalize BetweenArray function from R package “LIMMA” from the Bioconductor project.10 After normalization, the Student’s t-test was introduced to compare difference between the NAC-resistant group and the NAC-sensitive group. P-value <0.05 and |fold change (FC)|>2 were set as the thresholds for screening DE-miRNAs. This screening method was defined as “Previous analysis.” In addition, the online GEO2R analytic tool provided by the GEO database was further validated by our analytic results of “Previous analysis.”
Prediction of target genes of DE-miRNAs
The miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php), an experimentally validated microRNA-target interactions database, is widely used for predicting target genes of miRNA.11 In this study, miRTarBase was applied to predict targets of the top 10 most upregulated and downregulated DE-miRNAs.
Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis
GO annotation and KEGG pathway enrichment analyses for the predicted target genes of the selected 20 DE miRNAs were conducted by using the online website, the database for annotation, visualization, and integrated discovery (DAVID, http://david-d.ncifcrf.gov/).12
Construction and analysis of PPI network and miRNA-target network
To assess the interactive relationship among these predicted target genes of the top10 most upregulated and downregulated DE-miRNAs, the STRING database (http://string-db.org) was introduced to construct the PPI network.13 In search of hub genes in the two networks, Cytoscape software (version 3.6.1) was used to analyze the degree of connectivity.14 According to degree, we screened out the top 25 hub genes, after which miRNA-hub genes networks were established by Cytoscape software.
Cell culture
The human breast cancer cell line, BCap37, was purchased from the cell bank of the Chinese Scientific Academy. Two multidrug-resistant breast cancer cells, Bads-200 and Bats-72, were previously selected by our lab via treatment of Bcap37 for dose-dense and dose-escalation administration with paclitaxel.15 The three breast cancer cell lines, Bcap37, Bads-200, and Bats-72, were maintained in RMPI-1640 medium (Gibco, Life Technologies, Carlsbad, CA, USA) with 10% FBS Biological Industries, Cormwell, CT, USA) and 1% penicillin-streptomycin at 37°C and 5% CO2.
RNA extraction and quantitative-PCR (q-PCR)
Total RNA was extracted from Bcap37, Bads-200, and Bats-72 cell lines by using RNAiso plus Reagent (TaKaRa, Kusatsu, Japan). MiRNAs reverse transcription primers and q-PCR primers were purchased from Ribobio Co. Ltd (Guangzhou, China). Total RNA was first reversely transcribed into complementary DNA through the PrimeScript RT Reagent Kit (TaKaRa, RR0037A). Then, SYBR Premix Ex Taq (TaKaRa, RR420A) was employed to conduct q-PCR. All experiments were performed in triplicate. The U6 small nuclear RNA was used as the internal control.
Expression data from the cancer genome atlas (TCGA)
The miRNA-sequencing level three data in patients of breast invasive carcinoma was downloaded from TCGA database (https://genome-cancer.ucsc.edu/). The expression levels of potential miRNAs were extracted from the downloaded miRNA information. Statistical analysis between two groups was performed using Student’s t-test. Differences were considered significant with a P-value <0.05.
Drug sensitivity test
The roles of miR-3617-3p, miR-3136-3p, and miR-520b in modulating breast cancer chemoresistance were determined using drug sensitivity test, as reported previously.9 In total, 2,500 cells of Bads-200 and Bats-72 were seeded in 96-well plates and were cultured for 12 hours, after which cells were transfected with miRNA mimics or negative control miRNA (Ribobio Co. Ltd) at a final concentration of 50 nM using LipofectamineTM 3000 (Invitrogen, Carlsbad, CA, USA). After 24 hours, the cells were treated with paclitaxel for 48 hours. Then, MTT solution was added and incubated for 4 hours. Subsequently, the medium was replaced with 0.15 mL dulbecco’s modified eagle media. Finally, the absorbance in each well was detected at 570 nm through a microplate reader (Bio-Rad, Sunnyvale, CA, USA).
Results
Identification of DE-miRNAs and their targets in chemoresistant breast cancer
A microarray data set GSE73736 was downloaded from the GEO database. This data set included 10 cases of chemoresistant breast cancer tissues (GSM1901882–GSM1901891) and 10 cases of chemosensitive tissues (GSM1901892–GSM1901901). The data were first normalized, and the results are presented in Figure 1. Then, the data were further processed by unpaired t-test (P<0.05, |log2FC|>1). In total, 123 DE-miRNAs were successfully screened out, as shown in Figure 2A. Among the 123 DE-miRNAs, 60 miRNAs were significantly upregulated and 63 miRNAs were downregulated. The top 10 most upregulated and downregulated miRNAs are listed in Tables 1 and 2, respectively.
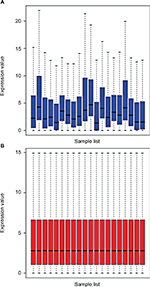  | Figure 1 Normalization of GSE73736. (A) Data before standardization; (B) data after standardization. |
  | Table 1 Top 10 most upregulated differentially expressed miRNAs between DR group and DS group of human breast cancer Abbreviations: DR, drug resistant; DS, drug sensitive; FC, fold change. |
  | Table 2 Top 10 most downregulated differentially expressed miRNAs between DR group and DS group of human breast cancer Abbreviations: DR, drug resistant; DS, drug sensitive; FC, fold change. |
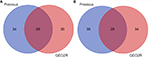
In order to determine the reliability of our screening method, we further analyzed DE-miRNAs using the online analytic tool, GEO2R, provided by GEO database. Finally, 118 significant DE-miRNAs (P<0.05), including 59 upregulated miRNAs and 59 downregulated miRNAs, were obtained. Through getting intersection of the two sets of DE-miRNAs, we discovered that a lot of miRNAs commonly appeared in both two sets, as shown in Figure S1.
Next, miRTarbase, an experimentally validated miRNA-target interactions database, was employed to predict the target genes of the top 10 most upregulated and downregulated DE-miRNAs. Finally, 1,763 potential target genes were generated, among which 617 were for the upregulated miRNAs and 1,146 for the downregulated miRNAs.
Functional and pathway enrichment analyses
To further explore the systematic features and biological functions of the aforementioned potential target genes, GO functional annotation and KEGG pathway enrichment analyses were performed via the online website, DAVID. Three GO categories, including BP, cellular component (CC), and molecular function (MF), were selected in the functional annotation.
The top 10 GO terms of the target genes of the top 10 most upregulated DE-miRNAs are shown in Figures 3A–C, including regulation of transcription and regulation of anti-apoptosis in the BP category, nuclear lumen and intracellular organelle lumen in the CC category, and DNA binding and zinc ion binding in the MF category. KEGG pathway analysis (Figure 3D) for the target genes of the top 10 most upregulated DE-miRNAs indicated that these target genes were enriched in some types of cancer, including glioma, melanoma, chronic myeloid leukemia, and non-small cell lung cancer. In addition, some of these potential target genes were also related to other pathways, such as the insulin signaling pathway and focal adhesion.
Subsequently, we conducted GO functional and KEGG pathway analyses for the target genes of the top 10 most downregulated DE-miRNAs. These target genes were involved in a variety of GO terms, such as regulation of transcription, negative regulation of apoptosis, and negative regulation of programmed cell death in the BP category (Figure 4A), intracellular organelle lumen and membrane-enclosed lumen in the CC category (Figure 4B), and ion binding and nucleotide binding in the MF category (Figure 4C). The enriched KEGG pathways (Figure 4D) for these target genes included the MAPK signaling pathway, wnt signaling pathway, and p53 signaling pathway. Of note, the three signaling pathways have been demonstrated to be associated with chemoresistance in the course of cancer treatment.9,16,17 Besides, 34 target genes (TRAF1, FGF7, XIAP, FOXO1, BCL2L1, PTEN, IGF1R, RAC1, FAS, RUNX1, TRAF6, MYC, FZD9, EGFR, CTBP1, MSH3, TGFBR2, CYCS, TP53, ITGA2, CDK6, CDK4, STAT3, RAD51, FZD6, MAPK1, CCDC6, NRAS, CDKN1A, LAMA4, CCND1, CRKL, LAMC1, and CRK) were involved in pathways in cancer.
Construction and analysis of PPI network and miRNA-target network
It has been widely acknowledged that proteins rarely act alone, but interact with each other. In view of this perspective, the STRING database was applied to construct the PPI network of these potential target genes of the top 10 most downregulated and upregulated miRNAs. After analyzing the data from STRING using Cytoscape software, we screened out the top 25 hub nodes according to degree (Table 3). For downregulated miRNAs, the top 10 hub genes were MAPK1, CALM3, ESR1, CCND1, PPP2CA, CDKN1A, CYCS, CREB1, CDC27, and IGF1R, among which MAPK1 showed the highest node degree (degree=57). For the upregulated miRNAs, the top 10 hub genes were PRDM10, PHLPP2, PIK3CB, H2AFX, CALM3, POLR2F, HSPA5, DICER1, ACTG1, and BTRC. Among these hub genes, PRDM10 showed the highest node degree, which was 67. For better visualization of interactions of these hub genes, additionally, we constructed networks based on the screened top 25 hub genes, as presented in Figure 5.
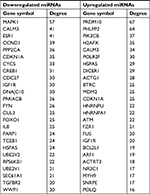  | Table 3 Hub genes identified in the PPI networks Abbreviation: PPI, protein–protein interaction. |
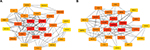  | Figure 5 The top 25 hub genes. (A) For downregulated miRNAs; (B) for upregulated miRNAs. |
Next, we further constructed the miRNA-hub gene network using Cytoscape software. As shown in Figure 6A, the hub target genes of upregulated miRNAs could be potentially regulated by the top 10 most upregulated miRNAs. Of note, miR-4766-3p and miR-4697-5p could regulate the most hub target genes (n=4), suggesting that the two miRNAs may possess a significant role in manipulating drug resistance of human breast cancer. The results presented in Figure 6B demonstrate that the top 25 hub genes from target genes of the top 10 most downregulated miRNAs could be potentially modulated by six miRNAs, namely miR-1245b-5p, miR-1253, miR-922, miR-520b, miR-584-5p, and miR-92a-1-5p. Among these miRNAs, miR-520b was found to potentially target the most hub genes (n=11). Additionally, miR-1253 and miR-584-5p could regulate 10 and six hub genes, respectively. The three miRNAs may act as crucial negative regulators in chemoresistance of breast cancer.
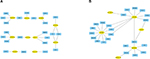  | Figure 6 MiRNA–hub gene networks. (A) For upregulated miRNAs; (B) for downregulated miRNAs. |
Altogether, the construction and analysis of the PPI network could provide us with some information in searching for key genes (such as MAPK1 and PRDM10) which play important roles in chemoresistance of breast cancer. The construction of the miRNA-hub gene network is considerably helpful for us to catch sight of the most potentially functional miRNAs.
Expression and roles of potential miRNAs in breast cancer
Two chemoresistant cell lines, Bads-200 and Bats-72, originated from chemosensitive Bcap37, were chosen to determine the expression of the 16 potential miRNAs mentioned above in chemoresistant breast cancer. As shown in Figure 7A, miR-3617-3p, miR-3136-3p, and miR-3130-5p were significantly upregulated, whereas miR-584-5p and miR-520b were downregulated in Bads-200 cell when compared to Bcap37. Expression levels of miR-3617-3p, miR-3136-3p, miR-4277, and miR-92a-1-5p in Bats-72 cell were obviously much higher than the Bcap37 cell, whereas miR-520b was significantly downregulated in the Bats-72 cell (Figure 7B). In conclusion, compared to the Bcap37 cell, the expression values of miR-3617-3p and miR-3136-3p were commonly increased, whereas miR-520b was decreased in Bads-200 and Bats-72 cell lines. Next, we further analyzed miR-3617-3p, miR-3136-3p, and miR-520b expression in breast cancer tissues by using the TCGA database, and eventually discovered that miR-3136-3p and miR-520b were, respectively, upregulated and downregulated in breast cancer tissues compared to adjacent normal breast tissues (Figure S2). However, miRNA miR-3617-3p was not found in the downloaded miRNA data from TCGA.
Next, we evaluated the roles of miR-3617-3p, miR-3136-3p, and miR-520b in modulation of breast cancer chemoresistance. Figures 7C and D demonstrated that miR-520b could effectively overcome paclitaxel resistance, while miR-3136-3p could significantly promote the development and progression of paclitaxel resistance in both Bads-200 and Bats-72 cells. For miR-3617-3p, we found that it could only reduce the chemosensitivity of the Bats-72 cell.
In a few words, all these findings suggested that miR-3617-3p, miR-3136-3p, and miR-520b may be three key regulators of chemoresistance in breast cancer.
Discussion
It has been widely acknowledged that miRNAs, a class of small endogenous noncoding RNAs, are frequently dysregulated in breast cancer. The abnormal expression of miRNAs is closely associated with the occurrence and development of breast cancer, such as proliferation,18 apoptosis,19 angiogenesis,20 and metastasis.21 Recently, our group has found that miR-20a and miR-27b were involved in chemoresistance of breast cancer.7,9 The roles of miRNAs in chemoresistance of breast cancer was also reported by other groups.8 However, studies regarding systematic analysis of miRNAs’ roles in breast cancer NAC resistance are still rare.
In this study, we first screened out 123 DE-miRNAs in chemoresistant breast cancer tissues compared to chemosensitive breast cancer tissues. Among these DE-miRNAs, miR-1253 and miR-1911-3p were shown to be the most downregulated and upregulated in the chemoresistance group. Previous investigations indicated that miR-1253 acted as a tumor suppressor in non-small cell lung cancer.22 MiR-1911-3p has been reported to have a correlation with neuroglioma.23 In addition to the two miRNAs, most identified DE-miRNAs have been confirmed to be associated with tumorigenesis.
KEGG pathway enrichment analysis revealed that the potential target genes of the top 10 most upregulated and downregulated DE-miRNAs were enriched in some cancer-associated pathways, such as glioma, melanoma, chronic myeloid leukemia, and non-small cell lung cancer, and some chemoresistance-associated pathways, such as the MAPK signaling pathway, wnt signaling pathway, and p53 signaling pathway.
In the PPI networks, MAPK1 and PRDM10 were the hub genes with the highest connectivity degree of 57 and 67, respectively. MAPK1 has been demonstrated to be involved in chemoresistance of breast cancer in our previous study.9 PRDM10, a member of PRDI-BF1 and RIZ homology domain containing (PRDM), has been lately discovered to be linked with the onset and development of several malignancies, such as nasopharygeal carcinoma24 and undifferentiated pleomorphic sarcoma.25 However, currently, studies about the impact of PRDM10 on chemoresistance are still absent. Therefore, the function and mechanism of PRDM10 in chemoresistance of breast cancer need to be further investigated.
Through constructing hub gene-miRNA networks, we found that the top 25 hub genes could be potentially modulated by 16 DE-miRNAs (from the top 10 most upregulated and downregulated DE-miRNAs). Among these 16 DE-miRNAs, miR-520b was the miRNA which could potentially regulate the most hub genes (n=11). Cui et al26 suggested that overexpression of miR-520b could increase the sensitivity of breast cancer cell lines to complement-dependent cytotoxicity. Additionally, miR-520b has also been reported to increase chemosensitivity of hepatocellular carcinoma cells to doxorubicin.27 All these findings support that miR-520b may be a key negative modulator in breast cancer NAC resistance.
Then, the expression values of these miRNAs in Bads-200, Bats-72, and Bcap37 were determined by q-PCR. When compared with Bcap37, miR-3617-3p, and miR-3136-3p were markedly upregulated and miR-520b was significantly downregulated in both Bads-200 and Bats-72 cells. In view of this, the three miRNAs may be the most important regulators in breast cancer NAC resistance.
Next, we further examined the role of miR-3617-3p, miR-3136-3p, and miR-520b in regulating breast cancer chemoresistance. Paclitaxel was chosen as the first tested drug as it is a chemotherapy medication employed to treat multiple types of cancer. In breast cancer, particularly, paclitaxel is considered as one of the first-line chemotherapeutic agents and is also widely used as an NAC agent clinically.28 Our results suggested that all the three miRNAs, especially miR-3136-3p and miR-520b, are key regulators in breast cancer chemoresistance.
Besides, miR-3130-5p and miR-584-5p were upregulated and downregulated in the Bads-200 cell. MiR-4277 and miR-92a-1-5p were upregulated in the Bats-72 cell. These miRNAs may also play important roles in modulating breast cancer NAC resistance. Despite some positive results being obtained in our current study, many more experiments need to be launched to confirm our preliminary analytic data in the future.
Conclusion
To sum up, our present study provides a comprehensive analysis of DE-miRNAs and their potential target genes which may be involved in breast cancer NAC resistance. All findings have demonstrated that miR-3617-3p, miR-3136-3p, and miR-520b may be three key functional miRNAs in regulating breast cancer NAC resistance, and they may serve as NAC response predictors, prognosticators, and therapeutic targets in the future.
Acknowledgments
This work was supported by National Natural Science Foundation of China grants (81372462, 81572987) and the Department of Science and Technology of Zhejiang Province (2014C03012). Thanks to the anonymous reviewers for their valuable comments and suggestions that helped improve the quality of our manuscript.
Author contributions
WYL, JXL, and WMF designed the study. WYL and JXL wrote the manuscript. WYL and BSD performed the experiments and statistical analysis of the data. LX polished the manuscript. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Desantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67(6):439–448. | ||
Kaufmann M, von Minckwitz G, Mamounas EP, et al. Recommendations from an international consensus conference on the current status and future of neoadjuvant systemic therapy in primary breast cancer. Ann Surg Oncol. 2012;19(5):1508–1516. | ||
Demichele A, Yee D, Esserman L. Mechanisms of resistance to neoadjuvant chemotherapy in breast cancer. N Engl J Med. 2017;377(23):2287–2289. | ||
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. | ||
Ambros V. The functions of animal microRNAs. Nature. 2004; 431(7006):350–355. | ||
Shi X, Zhang H, Wang M, et al. LncRNA AFAP1-AS1 promotes growth and metastasis of cholangiocarcinoma cells. Oncotarget. 2017;8(35):58394–58404. | ||
Chen D, Si W, Shen J, et al. miR-27b-3p inhibits proliferation and potentially reverses multi-chemoresistance by targeting CBLB/GRB2 in breast cancer cells. Cell Death Dis. 2018;9(2):188. | ||
Lyu H, Wang S, Huang J, Wang B, He Z, Liu B. Survivin-targeting miR-542-3p overcomes HER3 signaling-induced chemoresistance and enhances the antitumor activity of paclitaxel against HER2-overexpressing breast cancer. Cancer Lett. 2018;420:97–108. | ||
Si W, Shen J, du C, et al. A miR-20a/MAPK1/c-Myc regulatory feedback loop regulates breast carcinogenesis and chemoresistance. Cell Death Differ. 2018;25(2):406–420. | ||
Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21(9):2067–2075. | ||
Chou CH, Shrestha S, Yang CD, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. | ||
Huang Daw, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. | ||
Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. | ||
Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. | ||
Jiang D, Sui M, Zhong W, Huang Y, Fan W. Different administration strategies with paclitaxel induce distinct phenotypes of multidrug resistance in breast cancer cells. Cancer Lett. 2013;335(2):404–411. | ||
Yang W, Li Y, Song X, Xu J, Xie J. Genome-wide analysis of long noncoding RNA and mRNA co-expression profile in intrahepatic cholangiocarcinoma tissue by RNA sequencing. Oncotarget. 2017;8(16):26591–26599. | ||
Ozaki T, Yu M, Yin D, et al. Impact of RUNX2 on drug-resistant human pancreatic cancer cells with p53 mutations. BMC Cancer. 2018;18(1):309. | ||
Luo T, Yan Y, He Q, Ma X, Wang W. miR-328-5p inhibits MDA-MB-231 breast cancer cell proliferation by targeting RAGE. Oncol Rep. 2018;39(6):2906-2914. | ||
Zhao W, Zhang X, Zhou Z, et al. Liraglutide inhibits the proliferation and promotes the apoptosis of MCF-7 human breast cancer cells through downregulation of microRNA-27a expression. Mol Med Rep. 2018;17(4):5202–5212. | ||
Ozawa T, Matsuyama T, Toiyama Y, et al. CCAT1 and CCAT2 long noncoding RNAs, located within the 8q.24.21 ‘gene desert’, serve as important prognostic biomarkers in colorectal cancer. Ann Oncol. 2017;28(8):1882–1888. | ||
Cheng CW, Wang HW, Chang CW, et al. MicroRNA-30a inhibits cell migration and invasion by downregulating vimentin expression and is a potential prognostic marker in breast cancer. Breast Cancer Res Treat. 2012;134(3):1081–1093. | ||
Liu M, Zhang Y, Zhang J, et al. MicroRNA-1253 suppresses cell proliferation and invasion of non-small-cell lung carcinoma by targeting WNT5A. Cell Death Dis. 2018;9(2):189. | ||
Yang H, Wang Y. Five miRNAs considered as molecular targets for predicting neuroglioma. Tumour Biol. 2016;37(1):1051–1059. | ||
Zamanian Azodi M, Rezaei Tavirani M, Rezaei Tavirani M, Vafaee R, Rostami-Nejad M. Nasopharyngeal carcinoma protein interaction mapping analysis via proteomic approaches. Asian Pac J Cancer Prev. 2018;19(3):845–851. | ||
Hofvander J, Tayebwa J, Nilsson J, et al. Recurrent PRDM10 gene fusions in undifferentiated pleomorphic sarcoma. Clin Cancer Res. 2015;21(4):864–869. | ||
Cui W, Zhang Y, Hu N, et al. miRNA-520b and miR-520e sensitize breast cancer cells to complement attack via directly targeting 3′UTR of CD46. Cancer Biol Ther. 2010;10(3):232–241. | ||
Gao AM, Zhang XY, Hu JN, Ke ZP, Jn H, Zp K. Apigenin sensitizes hepatocellular carcinoma cells to doxorubic through regulating miR-520b/ATG7 axis. Chem Biol Interact. 2018;280:45–50. | ||
Rivera E, Gomez H. Chemotherapy resistance in metastatic breast cancer: the evolving role of ixabepilone. Breast Cancer Res. 2010;12(Suppl 2):S2. |
Supplementary materials
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.