Back to Journals » International Journal of General Medicine » Volume 14
Identification of ceRNA (lncRNA-miRNA-mRNA) Regulatory Network in Myocardial Fibrosis After Acute Myocardial Infarction
Authors Wang S, Liu Y, Hu X, Zhang X, Xu L, Yang Y, Wu R, Wang E, Lv T
Received 19 July 2021
Accepted for publication 2 December 2021
Published 19 December 2021 Volume 2021:14 Pages 9977—9990
DOI https://doi.org/10.2147/IJGM.S329391
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Shuo Wang, Yuying Liu, Xitian Hu, Xiaolei Zhang, Lei Xu, Yan Yang, Rubing Wu, Enmao Wang, Tianjie Lv
Department of Cardiovasology, Shijiazhuang People’s Hospital, Shijiazhuang, People’s Republic of China
Correspondence: Xitian Hu
Department of Cardiovasology, Shijiazhuang People’s Hospital, No. 9 Fangbei Road, Shijiazhuang, Hebei, 050000, People’s Republic of China
Tel +86-17603119015
Email [email protected]
Purpose: Myocardial fibrosis (MF) after acute myocardial infarction (AMI) ultimately results in heart failure, which is a serious threat to human life. This study aimed to excavate critical biomarkers associated with MF after AMI.
Materials and Methods: RNA-sequencing was performed to obtain differentially expressed mRNAs (DEmRNAs), miRNAs (DEmiRNAs) and lncRNAs (DElncRNAs) in AMI and MF after AMI.
Results: Abundant DEmRNAs, DEmiRNAs and DElncRNAs were identified in AMI and MF after AMI. The ceRNA network, which contained 9 lncRNA-miRNA pairs and 9 miRNA-mRNA pairs, was acquired. In AMI, all candidate markers generally exhibited the same pattern as that in our RNA-seq results; while in MF after AMI, except for CENPB, JAK2 and hsa-miR-197-3p, the expression of the others in the qRT-PCR results exhibited the same pattern as that in our RNA-seq results.
Conclusion: We speculated that LINC00664/hsa-miR-197-3p/JAK2 and GAS6-AS1/SNHG22/hsa-miR-135a-5p/CENPB/BCL9L interaction pairs may serve as potential biomarkers in MF after AMI.
Keywords: acute myocardial infarction, myocardial fibrosis, lncRNA, ceRNA network
Introduction
Acute myocardial infarction (AMI) is a life-threatening cardiovascular disease with high mortality and morbidity globally, featured by myocardial necrosis caused by acute or persistent ischemia and hypoxia of cardiac muscle.1 Myocardial fibrosis (MF) is a main pathological changes appear in the heart after AMI, characterized by increasing cardiac fibroblasts activity, increasing the risk of heart failure.2 Thus, preventing and reversing cardiac fibrosis may have potential therapeutic effects on improving heart function and even reducing mortality risk.
With the rapid development of RNA-sequencing technology, the roles of non-coding RNAs (ncRNAs) in cardiovascular disease has been extensively studied. Chistiakov et al have highlighted the important role of miRNA in cardiac function under physiological conditions and cardiac dysfunction under pathological conditions.3 Wang et al concluded the regulatory functions or diagnostic potential of miRNAs, siRNAs, lncRNAs and circRNAs in cardiovascular diseases.4 Similarly, a review indicated that ncRNAs, including lncRNAs, miRNAs and circRNAs, are emerging key regulators of gene expression of several physiological and pathological processes in AMI, as well as lncRNA/circRNA-miRNA-mediated interaction (ie, competitive endogenous RNA (ceRNA) mechanism).5 Thum reviewed the role of ncRNAs in MF and suggested that ncRNAs can serve as powerful therapeutic targets to treat MF.6 PLAUR was found to play a vital role in transducing inter-cellular signals from endothelial cells and fibroblast cells to intra-cellular pathways of myocardial cells, leading to gene expression involved in cellular response to hypoxia.7 Zhuo et al indicated that lncRNA SNHG8 may serve as diagnostic or prognostic biomarkers of AMI.8 It was reported that miR-223 enhanced cell proliferation, migration, and differentiation in cardiac fibrosis, thus mediated cardiac fibrosis after MI partially via the involvement of RASA1.9 Based on the above-mentioned findings, therefore, we performed this current work.
In current study, differentially expressed mRNAs (DEmRNAs), miRNAs (DEmiRNAs) and lncRNAs (DElncRNAs) in AMI and MF after AMI were acquired, respectively. Then, shared DEmRNAs/DEmiRNAs/DElncRNAs in AMI and MF after AMI were further determined. Finally, a ceRNA (DElncRNA-DEmiRNA-DEmRNA) regulatory network was constructed to explore the potential molecular mechanisms of MF after AMI.
Materials and Methods
Subjects and Samples
The cohort subjected to RNA-Seq comprised 3 patients with AMI, 3 patients with MF after AMI and 3 healthy individuals. The inclusion criteria of AMI patients were as follows: (1) time of chest pain or distress >30min within 24h, and the level of the myocardial enzyme of creatine kinase (CK)-MB and cardiac troponin T (cTnT) was higher than the normal range; (2) patients had their first episode; (3) patients received no medical or surgical treatment prior to admission; (4) patients had blood samples before admission, at discharge, and 6 months after MI; and (5) patients had complete clinical data. The exclusion criteria of AMI patients were as follows: (1) patients with myocarditis and other diseases caused by chest pain or distress; (2) patients with a history of renal failure, advanced liver disease, malignant tumors, and other inflammatory diseases such as psoriasis and rheumatoid arthritis; (3) recurrent patients; (4) patients with incomplete clinical data; and (5) patients with missing blood samples before admission, at discharge, and 6 months after MI. Those patients diagnosed with MF (1 year after MI) were enrolled. All samples were collected after obtaining written informed consent from every participant. This study was approved by the ethics committee of Shijiazhuang People’s Hospital and performed in accordance with the Declaration of Helsinki. Peripheral whole blood (2.5 mL) drawn from each subject was collected in PAXgene® RNA blood tubes. Specifically, the blood sample from patients with AMI was collected within 12 h after onset of AMI.
RNA Isolation and Sequencing
According to the manufacturer’s protocol, RNA was extracted with PAXgene blood RNA kit. With Agilent 2100 Bioanalyzer (Agilent RNA 6000 Nano Kit), the concentration, integrity and RNA integrity number (RIN) values of RNA were assessed. After total RNA DNase I treatment, removal of rRNA, RNA interruption, synthesis of reverse transcriptional one and two strands, end repair, cDNA with an “A” at the 3ʹ end, connection of cDNA 5ʹ adapter, digestion of two strands of cDNA, PCR reaction, and product recovery and library quality inspection, DNBSEQ platform (PE100 strategy) was used to perform RNA sequencing for mRNA and lncRNA. The Fastx-Toolkit was used to trim 5ʹ and 3ʹ segments of reads to remove bases with mass <20 and delete reads with N >10%. HISAT2 was applied to align the clean reads with the human reference genome (GRCh38). Expression of mRNAs and lncRNAs was normalized and outputted with StringTie. The Rfam was used for annotation analysis on measured small RNA. The mature miRNA and miRNA precursor sequences were downloaded from miRBase. The expression of miRNA was quantified with miRDeep2. Finally, a total of 19,951 mRNAs, 1313 miRNAs and 16,054 lncRNAs were obtained.
Identification of DEmRNAs, DEmiRNAs and DElncRNAs in AMI and MF After AMI
DEGseq2 (http://bioconductor.org/packages/DESeq2/) was applied to identify DEmRNAs, DEmiRNAs and DElncRNAs in AMI and MF after AMI with |log2FC| ≥ 1 and p-value < 0.05. Hierarchical clustering analysis of DEmRNAs, DEmiRNAs and DElncRNAs was performed with R (https://www.r-project.org/) package “pheatmap”. Then, shared DEmRNAs in AMI and MF after AMI were further identified with Venny 2.1.0 (https://bioinfogp.cnb.csic.es/tools/venny/). CPDB (http://cpdb.molgen.mpg.de/CPDB) was used to perform GO and KEGG enrichment analysis for up-regulated and down-regulated shared DEmRNAs in AMI and MF after AMI. A value of p < 0.05 was considered to be represented statistically significant.
CeRNA (DElncRNA-DEmiRNA-DEmRNA) Regulatory Network
Six bioinformatic algorithms (PITA, RNA22, miRmap, microT, miRanda and PicTar) were used to predict the targeted DEmRNAs of DEmiRNAs. Then, the DEmiRNA-DEmRNA pairs recorded by ≥4 algorithms were remained for network construction. DIANA-LncBase v2.0 was applied to predict DElncRNA-DEmiRNA interaction pairs. Based on ceRNA theory, the ceRNA (DElncRNA-DEmiRNA-DEmRNA) regulatory network was constructed with Cytoscape, by using a combination of lncRNA-miRNA pairs and miRNA-mRNA pairs.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Validation
Thirteen blood samples were obtained from 5 patients with AMI (3 for RNA sequence), 3 patients with MF after AMI (2 for RNA sequence) and 5 healthy controls (3 for RNA sequence). Total RNA was isolated with the Trizol reagent (Invitrogen, USA) following manufacturer’s protocol. The qRT-PCR reactions were performed in ABI 7300 Real-time PCR Detection System with SuperReal PreMix Plus (SYBR GREEN) (Invitrogen, USA). Relative gene expression was analyzed by 2−ΔΔCT method. The human GAPDH and ACTB were used as endogenous controls for mRNA and lncRNA expression in analysis. The human U6 was used as endogenous controls for miRNA in analysis. The PCR primers were displayed in Table S1.
Validation in the GEO Database
To further validate our findings, GSE66360, including miRNA and mRNA expression profiles and consisting of 49 patients experiencing acute myocardial infarction and 50 healthy cohorts, was downloaded from GEO database. Then, the expression levels of selected mRNAs and lncRNAs were validated with GSE66360.
Results
Clinical Characteristics of Enrolled Individuals for RNA Sequencing and qRT-PCR Validation
A total of 14 individuals (5 patients with AMI, 4 patients with MF after AMI, and 5 healthy controls) were enrolled in RNA sequencing and qRT-PCR validation. The clinical information of these individuals was presented in Table S2.
Identification of DEmRNAs, DEmiRNAs and DElncRNAs in AMI and MF After AMI
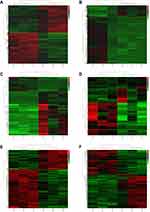
Compared to normal controls, 1990 DEmRNAs (1047 up- and 943 down-regulated) were identified in AMI, 1477 DEmRNAs (290 up- and 1187 down-regulated) were identified in MF after AMI, respectively (Figure 1A and B). Of these, PFKFB2 and SLC4A10, TVP23C-CDRT4 and UBAP1L was the most up- and down-regulated DEmRNA in AMI and MF after AMI, respectively (Table 1). A total of 509 shared DEmRNAs (84 up- and 425 down-regulated) in AMI and MF after AMI were acquired (Figure S1A).
 |
Table 1 Top 10 Up- and Down-Regulated DEmRNAs |
 |
Figure 2 Continue. |
Compared to normal controls, 88 DEmiRNAs (22 up- and 66 down-regulated) were identified in AMI, 36 DEmiRNAs (16 up- and 20 down-regulated) were identified in MF after AMI, respectively (Figure 1C and D). Of these, hsa-miR-223-3p and hsa-miR-6802-3p, hsa-miR-1299 and hsa-miR-197-3p was the most up- and down-regulated DEmiRNA in AMI and MF after AMI, respectively (Table 2). A total of 22 shared DEmiRNAs (6 up- and 16 down-regulated) in AMI and MF after AMI were acquired (Figure S1B).
 |
Table 2 Top 10 Up- and Down-Regulated DEmiRNAs |
Compared to normal controls, 473 DElncRNAs (315 up- and 158 down-regulated) were identified in AMI, 502 DElncRNAs (212 up- and 290 down-regulated) were identified in MF after AMI, respectively (Figure 1E and F). Of these, DLEU1 and FP236383.3, SBF2-AS1 and CTBP1-DT was the most up- and down-regulated DElncRNA in AMI and MF after AMI, respectively (Table 3). A total of 90 shared DElncRNAs (39 up- and 51 down-regulated) in AMI and MF after AMI were acquired (Figure S1C).
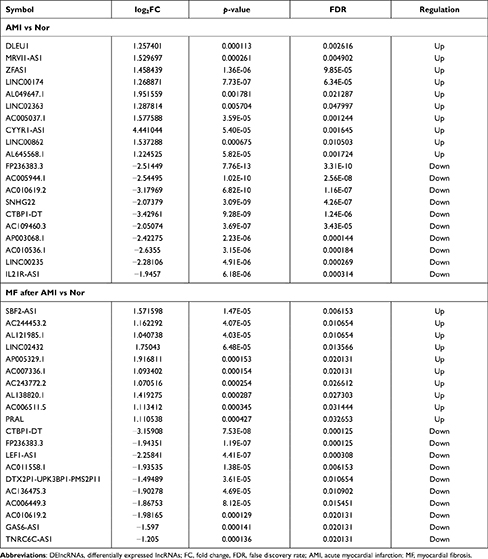 |
Table 3 Top 10 Up- and Down-Regulated DElncRNAs |
Functional Annotation of Shared DEmRNAs in AMI and MF After AMI
For up-regulated shared DEmRNAs group, the result indicated that these genes were enriched in response to stress (p = 9.94E-06), cell activation (p = 1.25E-04), enzyme regulator activity (p = 2.19E-04), Pantothenate and CoA biosynthesis (p = 3.53E-03) and Leishmaniasis (p = 4.09E-03) (Figure 2A–D). These, down-regulated genes were enriched in lymphocyte costimulation (p = 2.45E-05), leukocyte activation (p = 1.28E-04), protein binding (p = 3.69E-05), Th17 cell differentiation (p = 1.85E-03), NF-kappa B signaling pathway (p = 5.95E-03) (Figure 2E–H).
CeRNA (DElncRNA-DEmiRNA-DEmRNA) Regulatory Network
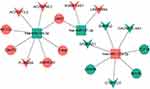
A total of 509 shared DEmRNAs, 22 shared DEmiRNAs and 90 shared DElncRNAs in AMI and MF after AMI were used to construct the ceRNA regulatory network. By overlapping DElncRNA-DEmiRNA interaction pairs and DEmiRNA-DEmRNA interaction pairs, a ceRNA network was acquired. The ceRNA network contained 9 lncRNA-miRNA pairs and 9 miRNA-mRNA pairs, including 9 lncRNAs, 3 miRNAs, and 8 mRNAs (Figure 3).
QRT-PCR Validation and Validation in GEO Database
Three DEmRNAs (BCL9L, CENPB and JAK2), three DEmiRNAs (hsa-miR-135a-5p, hsa-miR-144-3p and hsa-miR-197-3p) and two DElncRNAs (IL21R-AS1 and LEF1-AS1) were selected randomly to use for qRT-PCR validation. Based on our RNA-seq results, JAK2 and hsa-miR-135a-5p were up-regulated while BCL9L, CENPB, hsa-miR-144-3p, hsa-miR-197-3p, IL21R-AS1 and LEF1-AS1 were down-regulated in AMI and MF after AMI. In AMI, all these candidate markers generally exhibited the same pattern as that in our RNA-seq results; while in MF after AMI, except for CENPB, JAK2 and hsa-miR-197-3p, the expression of the others in the qRT-PCR results exhibited the same pattern as that in our RNA-seq results (Figure 4). As shown in Figure 5, the expression of BCL9L, CENPB, LEF1-AS1, LINC00664 and SNHG22 were found to exhibit the same pattern as that in our RNA-seq results.
Discussion
MF is a common hallmark in various heart diseases and predominant cause of morbidity and mortality in the world. The potential biomarkers for cardiovascular disease have been explored widely. It has been reported that ADMA is an independent predictor of cardiovascular morbidity and mortality.10,11 Zinellu et al suggested that plasma ADMA was involved in carotid narrowing after carotid endarterectomy intervention.12 Bivona et al indicated that serum Gal 3 levels rise immediately after AMI and decrease significantly within 5 days after the acute event.13 In addition, it has been concluded that H-FABP is not a reliable marker for AMI diagnosis.14 Despite available drug treatment strategy for MF, there is still an urgent need for development of new biomarkers to prevent and reverse MF after AMI.
Many lncRNAs have been reported to be involved in the development of various diseases. In this study, abundant lncRNAs were identified to be associated with AMI and MF after AMI, such as IL21R-AS1 and LEF1-AS1. Cai et al indicated that IL21R-AS1 was a candidate biomarker for coronary artery disease.15 Overexpressed LEF1-AS1 was detected in coronary artery atherosclerosis, which regulated vascular smooth muscle cell proliferation and migration.16 In agreement with previous studies, we observed IL21R-AS1 and LEF1-AS1 were significantly dyregulated and validated in the qRT-PCR as well in this analysis, which reminds us to focus on the roles of IL21R-AS1 and LEF1-AS1 in AMI and MF after AMI.
High LINC00664 expression was associated with shorter survival in endometrial and clear cell carcinomas patients, whereas, it is converse for high-grade serous carcinomas patients.17 Zhang et al determined an eight-lncRNA signature with high prognostic value for colorectal cancer, including LINC00664.18 Chen et al indicated LINC00664 was a prognostic marker for ovarian carcinoma.19 Chouvarine et al suggested that hypoxia alone could drive cardiac inflammation and identified miR-197-3p as a hypoxia-regulated miRNA.20 McManus et al examined associations of miRNA levels with 6 cardiometabolic (CM) traits and determined that miR-197-3p was significantly associated with body mass index, triglycerides, and systolic and diastolic blood pressures.21 The role of JAK/STAT signaling pathway in the heart has been extensively studied. Negoro et al demonstrated that JAK/STAT signaling pathway was activated in the rat model of AMI.22 Mascareno et al suggested that activation of the JAK/STAT signaling pathway contributed to the pathogenesis of myocardial ischemia.23 Chen et al indicated that activation of JAK2/STAT3 pathway can prevent apoptosis after AMI.24 Zhang et al revealed inhibition of JAK2/STAT3 pathway can attenuate myocardial hypertrophy induced by transverse aortic constriction.25 Chen et al implied that JAK/STAT pathway made contribution to left-atrial-fibrogenesis.26 Eid et al reported that activation of JAK2/STAT3 signaling contributed to reduce MI-induced left ventricle injury.27 In this study, JAK2 was a target of hsa-miR-197-3p, and LINC00664 may function as a ceRNA to capture hsa-miR-197-3p.
Altered expression of GAS6-AS1 has been correlated with poor outcome in various tumors. Han et al indicated that decreased GAS6-AS1 was related to poor prognosis in non-small cell lung cancer patients.28 Zhang et al demonstrated that high GAS6-AS1 expression was associated with advanced TNM stage in gastric cancer.29 Ai et al suggested that up-regulated GAS6-AS1 was negatively correlated with the overall survival of hepatocellular carcinoma patients.30 Similarly, SNHG22, located on chromosome 18q21.1, has been associated with multiple cancers as well. Zhang et al indicted that high expressed SNHG22 predicted poor prognosis in patients with epithelial ovarian carcinoma.31 Fang et al found that SNHG22 exerted carcinogenic effects in triple-negative breast cancer.32 Gao et al suggested that SNHG22 overexpression was correlated with worse overall survival in papillary thyroid cancer patients.33 However, to our best knowledge, the role of GAS6-AS1 and SNHG22 in the heart has not been reported yet.
Recent studies indicated that miR-135a-5p may play important roles in a variety of tumors, including gliomas, lung cancer and head and neck squamous cell carcinoma.34–36 In addition, Wu et al suggested that miR-135a inhibited cardiac fibrosis and it may be a therapeutic target for the amelioration of cardiac fibrosis.37 Xu et al demonstrated that miR-135a-5p negatively regulated hypoxia-induced cardiomyocyte injury in H9c2 cells.38 CENPB, a cell cycle-related gene, was reported to be differentially regulated in the heart tissue in serum-response factor null embryonic stem cells.39 BCL9L is a nuclear co-activator in the Wnt/β-catenin pathway, which was shown to be associated with various cancers. BCL9L knockdown was reported to inhibit pancreatic cancer progression and liver metastasis.40 Huge et al indicated that high expression of BCL9L was associated with poor overall survival in hepatocellular carcinoma patients.41 In addition, Cantù et al suggested a causative link between mutations in BCL9L and human congenital heart disease.42 Both CENPB and BCL9L were targeted by hsa-miR-135a-5p in this analysis. We speculated that GAS6-AS1 and SNHG22 may participate in development of MF after AMI via acting as ceRNAs of CENPB and BCL9L by sponging hsa-miR-135a-5p.
Conclusions
In conclusion, we speculated that LINC00664/hsa-miR-197-3p/JAK2 and GAS6-AS1/SNHG22/hsa-miR-135a-5p/CENPB/BCL9L interaction pairs may serve as potential biomarkers in MF after AMI. Due to small sample size included in this study, further research with a larger cohort is essential to be performed to confirm the conclusion. In addition, the specific mechanism of the lncRNA-miRNA-mRNA axes in MF after AMI need to be validated in vivo and in vitro.
Disclosure
The authors declare that there is no conflict of interest.
References
1. Virani SS, Alonso A, Benjamin EJ, et al. Heart disease and stroke statistics-2020 update: a report from the American heart association. Circulation. 2020;141(9):e139–e596. doi:10.1161/CIR.0000000000000757
2. Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-β1. Biochem Biophys Res Commun. 2017;487(4):769–775. doi:10.1016/j.bbrc.2017.04.044
3. Chistiakov DA, Orekhov AN, Bobryshev YV. Cardiac-specific miRNA in cardiogenesis, heart function, and cardiac pathology (with focus on myocardial infarction). J Mol Cell Cardiol. 2016;94:107–121. doi:10.1016/j.yjmcc.2016.03.015
4. Wang C, Jing Q. Non-coding RNAs as biomarkers for acute myocardial infarction. Acta Pharmacol Sin. 2018;39(7):1110–1119. doi:10.1038/aps.2017.205
5. Guo Y, Luo F, Liu Q, Xu D. Regulatory non-coding RNAs in acute myocardial infarction. J Cell Mol Med. 2017;21(5):1013–1023. doi:10.1111/jcmm.13032
6. Thum T. Noncoding RNAs and myocardial fibrosis. Nat Rev Cardiol. 2014;11(11):655–663. doi:10.1038/nrcardio.2014.125
7. Luo J, Wu L, Liu D, et al. Gene regulatory network analysis identifies key genes and regulatory mechanisms involved in acute myocardial infarction using bulk and single cell RNA-seq data. Math Biosci Eng. 2021;18(6):7774–7789. doi:10.3934/mbe.2021386
8. Zhuo LA, Wen YT, Wang Y, et al. LncRNA SNHG8 is identified as a key regulator of acute myocardial infarction by RNA-seq analysis. Lipids Health Dis. 2019;18(1):201. doi:10.1186/s12944-019-1142-0
9. Liu X, Xu Y, Deng Y, Li H. MicroRNA-223 regulates cardiac fibrosis after myocardial infarction by targeting RASA1. Cell Physiol Biochem. 2018;46(4):1439–1454. doi:10.1159/000489185
10. Zoccali C, Bode-Böger S, Mallamaci F, et al. Plasma concentration of asymmetrical dimethylarginine and mortality in patients with end-stage renal disease: a prospective study. Lancet. 2001;358(9299):2113–2117. doi:10.1016/S0140-6736(01)07217-8
11. Valkonen VP, Päivä H, Salonen JT, et al. Risk of acute coronary events and serum concentration of asymmetrical dimethylarginine. Lancet. 2001;358(9299):2127–2128. doi:10.1016/S0140-6736(01)07184-7
12. Zinellu A, Sotgia S, Porcu P, et al. Carotid restenosis is associated with plasma ADMA concentrations in carotid endarterectomy patients. Clin Chem Lab Med. 2011;49(5):897–901. doi:10.1515/CCLM.2011.121
13. Bivona G, Bellia C, Lo Sasso B, et al. Short-term changes in Gal 3 circulating levels after acute myocardial infarction. Arch Med Res. 2016;47(7):521–525. doi:10.1016/j.arcmed.2016.12.009
14. Bivona G, Agnello L, Bellia C, Lo Sasso B, Ciaccio M. Diagnostic and prognostic value of H-FABP in acute coronary syndrome: still evidence to bring. Clin Biochem. 2018;58:1–4. doi:10.1016/j.clinbiochem.2018.04.021
15. Cai Y, Yang Y, Chen X, et al. Circulating ‘lncRNA OTTHUMT00000387022ʹ from monocytes as a novel biomarker for coronary artery disease. Cardiovasc Res. 2016;112(3):714–724. doi:10.1093/cvr/cvw022
16. Zhang L, Zhou C, Qin Q, Liu Z, Li P. LncRNA LEF1-AS1 regulates the migration and proliferation of vascular smooth muscle cells by targeting miR-544a/PTEN axis. J Cell Biochem. 2019;120(9):14670–14678. doi:10.1002/jcb.28728
17. Fridley BL, Dai J, Raghavan R, et al. Transcriptomic characterization of endometrioid, clear cell, and high-grade serous epithelial ovarian carcinoma. Cancer Epidemiol Biomarkers Prev. 2018;27(9):1101–1109. doi:10.1158/1055-9965.EPI-17-0728
18. Zhang L, Chen S, Wang B, et al. An eight-long noncoding RNA expression signature for colorectal cancer patients’ prognosis. J Cell Biochem. 2019;120(4):5636–5643. doi:10.1002/jcb.27847
19. Chen Y, Bi F, An Y, Yang Q. Identification of pathological grade and prognosis-associated lncRNA for ovarian cancer. J Cell Biochem. 2019;120(9):14444–14454. doi:10.1002/jcb.28704
20. Chouvarine P, Legchenko E, Geldner J, Riehle C, Hansmann G. Hypoxia drives cardiac miRNAs and inflammation in the right and left ventricle. J Mol Med. 2019;97(10):1427–1438. doi:10.1007/s00109-019-01817-6
21. McManus DD, Rong J, Huan T, et al. Messenger RNA and MicroRNA transcriptomic signatures of cardiometabolic risk factors. BMC Genom. 2017;18(1):139. doi:10.1186/s12864-017-3533-9
22. Negoro S, Kunisada K, Tone E, et al. Activation of JAK/STAT pathway transduces cytoprotective signal in rat acute myocardial infarction. Cardiovasc Res. 2000;47(4):797–805. doi:10.1016/S0008-6363(00)00138-3
23. Mascareno E, El-Shafei M, Maulik N, et al. JAK/STAT signaling is associated with cardiac dysfunction during ischemia and reperfusion. Circulation. 2001;104(3):325–329. doi:10.1161/01.CIR.104.3.325
24. Chen H, Chen Y, Wang X, Yang J, Huang C. Edaravone attenuates myocyte apoptosis through the JAK2/STAT3 pathway in acute myocardial infarction. Free Radic Res. 2020;54(5):351–359. doi:10.1080/10715762.2020.1772469
25. Zhang Y, Zhang L, Fan X, et al. Captopril attenuates TAC-induced heart failure via inhibiting Wnt3a/β-catenin and Jak2/Stat3 pathways. Biomed Pharmacother. 2019;113:108780. doi:10.1016/j.biopha.2019.108780
26. Chen Y, Surinkaew S, Naud P, et al. JAK-STAT signalling and the atrial fibrillation promoting fibrotic substrate. Cardiovasc Res. 2017;113(3):310–320. doi:10.1093/cvr/cvx004
27. Eid RA, Alkhateeb MA, Eleawa S, et al. Cardioprotective effect of ghrelin against myocardial infarction-induced left ventricular injury via inhibition of SOCS3 and activation of JAK2/STAT3 signaling. Basic Res Cardiol. 2018;113(2):13. doi:10.1007/s00395-018-0671-4
28. Han L, Kong R, Yin DD, et al. Low expression of long noncoding RNA GAS6-AS1 predicts a poor prognosis in patients with NSCLC. Med Oncol. 2013;30(4):694. doi:10.1007/s12032-013-0694-5
29. Zhang P, Dong Q, Zhu H, Li S, Shi L, Chen X. Long non-coding antisense RNA GAS6-AS1 supports gastric cancer progression via increasing GAS6 expression. Gene. 2019;696:1–9. doi:10.1016/j.gene.2018.12.079
30. Ai J, Sun J, Zhou G, Zhu T, Jing L. Long non-coding RNA GAS6-AS1 acts as a ceRNA for microRNA-585, thereby increasing EIF5A2 expression and facilitating hepatocellular carcinoma oncogenicity. Cell Cycle. 2020;19(7):742–757. doi:10.1080/15384101.2020.1729323
31. Zhang PF, Wu J, Luo JH, et al. SNHG22 overexpression indicates poor prognosis and induces chemotherapy resistance via the miR-2467/Gal-1 signaling pathway in epithelial ovarian carcinoma. Aging. 2019;11(19):8204–8216. doi:10.18632/aging.102313
32. Fang X, Zhang J, Li C, Liu J, Shi Z, Zhou P. Long non-coding RNA SNHG22 facilitates the malignant phenotypes in triple-negative breast cancer via sponging miR-324-3p and upregulating SUDS3. Cancer Cell Int. 2020;20:252. doi:10.1186/s12935-020-01321-9
33. Gao H, Sun X, Wang H, Zheng Y. Long noncoding RNA SNHG22 increases ZEB1 expression via competitive binding with microRNA-429 to promote the malignant development of papillary thyroid cancer. Cell Cycle. 2020;19(10):1186–1199. doi:10.1080/15384101.2020.1749466
34. Guo LM, Ding GF, Xu W, et al. MiR-135a-5p represses proliferation of HNSCC by targeting HOXA10. Cancer Biol Ther. 2018;19(11):973–983. doi:10.1080/15384047.2018.1450112
35. Luo W, Sun C, Zhou J, et al. miR-135a-5p functions as a glioma proliferation suppressor by targeting tumor necrosis factor receptor-associated factor 5 and predicts patients’ prognosis. Am J Pathol. 2019;189(1):162–176. doi:10.1016/j.ajpath.2018.08.019
36. Zhang Y, Jiang WL, Yang JY, et al. Downregulation of lysyl oxidase-like 4 LOXL4 by miR-135a-5p promotes lung cancer progression in vitro and in vivo. J Cell Physiol. 2019;234(10):18679–18687. doi:10.1002/jcp.28508
37. Wu Y, Liu Y, Pan Y, et al. MicroRNA-135a inhibits cardiac fibrosis induced by isoproterenol via TRPM7 channel. Biomed Pharmacother. 2018;104:252–260. doi:10.1016/j.biopha.2018.04.157
38. Xu JJ, Zheng WH, Wang J, Chen YY. Long non-coding RNA plasmacytoma variant translocation 1 linked to hypoxia-induced cardiomyocyte injury of H9c2 cells by targeting miR-135a-5p/forkhead box O1 axis. Chin Med J. 2020;133(24):2953–2962. doi:10.1097/CM9.0000000000001147
39. Zhang SX, Garcia-Gras E, Wycuff DR, et al. Identification of direct serum-response factor gene targets during Me2SO-induced P19 cardiac cell differentiation. J Biol Chem. 2005;280(19):19115–19126. doi:10.1074/jbc.M413793200
40. Sannino G, Armbruster N, Bodenhöfer M, et al. Role of BCL9L in transforming growth factor-β (TGF-β)-induced epithelial-to-mesenchymal-transition (EMT) and metastasis of pancreatic cancer. Oncotarget. 2016;7(45):73725–73738. doi:10.18632/oncotarget.12455
41. Huge N, Sandbothe M, Schröder AK, et al. Wnt status-dependent oncogenic role of BCL9 and BCL9L in hepatocellular carcinoma. Hepatol Int. 2020;14(3):373–384. doi:10.1007/s12072-019-09977-w
42. Cantù C, Felker A, Zimmerli D, et al. Mutations in Bcl9 and Pygo genes cause congenital heart defects by tissue-specific perturbation of Wnt/β-catenin signaling. Genes Dev. 2018;32(21–22):1443–1458. doi:10.1101/gad.315531.118
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.