Back to Journals » Drug Design, Development and Therapy » Volume 15
Identification of Active Compounds and Mechanism of Huangtu Decoction for the Treatment of Ulcerative Colitis by Network Pharmacology Combined with Experimental Verification
Authors Chen W , He L, Zhong L, Sun J, Zhang L, Wei D, Wu C
Received 8 July 2021
Accepted for publication 17 September 2021
Published 29 September 2021 Volume 2021:15 Pages 4125—4140
DOI https://doi.org/10.2147/DDDT.S328333
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tuo Deng
Wenwen Chen,1,2,* Lin He,1,* Lian Zhong,1 Jiayi Sun,3 Lilin Zhang,1 Daneng Wei,1 Chunjie Wu1
1School of Pharmacy, Chengdu University of Traditional Chinese Medicine, Chengdu, 611137, People’s Republic of China; 2Department of Pharmacy, Chengdu Women’s and Children’s Central Hospital, School of Medicine, University of Electronic Science and Technology of China, Chengdu, 610091, People’s Republic of China; 3Innovative Institute of Chinese Medicine and Pharmacy, Chengdu University of Traditional Chinese Medicine, Chengdu, 611137, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Daneng Wei; Chunjie Wu Email [email protected]; [email protected]
Introduction: Huangtu decoction (HTD) has been widely used in the treatment of gastrointestinal bleeding, ulcerative colitis (UC) and gastrointestinal tumors in China, but its active compounds and mechanism are still not clear yet. The present research aimed to identify the active compounds and mechanism of HTD for the treatment of UC.
Methods: Firstly, the chemical compounds of HTD were qualitatively identified based on Q Exactive Orbitrap LC-MS/MS, and their potential targets were predicted through SwissTargetPrediction. Secondly, the differential expressed genes (DEGs) in colon tissues of UC patients and normal controls were retrieved from the GEO database. Thirdly, the overlapping targets of DEGs and the predicted targets were obtained and subjected to GO and KEGG analysis. Finally, the key targets in the most significantly enriched pathway were verified by in vivo experiment, and the protein and mRNA expressions of matrix metalloproteinase-1 (MMP1), MMP3, MMP7, MMP9 and MMP12 were determined by immunohistochemistry (IHC), Western blotting (WB) and quantitative real-time-PCR (qRT-PCR).
Results: A total of 47 compounds were identified and 29 overlapping targets were obtained from HTD extract. The most significantly enriched pathway of overlapping targets involved was MMP. HTD improved the pathological damage in colon tissues of DSS-induced UC model and significantly decreased the serum levels of IL-1β and IL-6. The protein and mRNA expressions of MMP1, MMP3 and MMP9 in colon tissues were significantly decreased after HTD treatment.
Conclusion: HTD treatment can alleviate the colonic inflammation via inhibiting MMPs including MMP1, MMP3 and MMP9.
Keywords: bioinformatic analysis, ulcerative colitis, Huangtu decoction, matrix metalloproteinases
Introduction
Ulcerative colitis (UC) is a kind of chronic colitis involving colorectal mucosa and submucosa, which is mainly caused by chronic inflammation and ulceration. The incidence of UC is increasing worldwide,1,2 and its clinical manifestations are primarily diarrhea, mucous purulent and bloody stool with abdominal pain and acute severe symptoms. It is estimated that the total annual economic burden of UC ranges from $8.1 to $14.9 billion in the United States and from €12.5 to €29.1 billion in Europe.3 The duration and extensive feature of disease are considered as important factors associated with an increased risk of colorectal cancer in patients with UC.4–6 Intense efforts have been made to elucidate the pathogeny of UC, but its molecular mechanisms are still poorly understood. Drug treatment is the first choice for UC, and the available drugs include mesalazine, corticosteroids, immunosuppressive drugs, and monoclonal antibodies to TNF-α currently in Western world.6 Nevertheless, the limitations of these drugs are also obvious, such as poor therapeutic effects, high price and bothersome side effects.7
In recent years, Traditional Chinese Medicine (TCM) has been gradually recognized as an effective method for treating various diseases not only in China and Southeast Asia, but also in many Western countries.8,9 TCM also plays an important role in drug development, providing abundant novel compound lead sources. Huangtu Decoction (HTD) was first recorded in the Treatise on “JingGuiYaoLue (Synopsis of Golden Chamber)” written around 205 A.D by Zhang Zhongjing, a famous medical scientist in the Eastern Han Dynasty. Its prescription is composed of 7 medicinal herbs, including Terra Flava Usta, Rehmanniae Radix, Scutellariae Radix, Aconiti Lateralis Radix Praeparata, Asini Corii Colla, Atractylodis Macrocephalae Rhizoma and Glycyrrhizae Radix et Rhizoma. HTD has been mainly used in the treatment of bleeding symptoms, such as blood under stool, hematemesis, bleeding, metrorrhagia and dull blood color, and highly respected by doctors for thousands of years in China.10,11 In modern times, HTD is often used to treat gastrointestinal bleeding, chronic UC, gastrointestinal tumor and so on.12,13 However, the active compounds and mechanism of HTD for the treatment of UC are still not clear.
During the last decades, bioinformatics analysis was widely used to screen genetic alterations at the genome level,14,15 so as to identify the differentially expressed genes (DEGs). Besides, network pharmacology has been successfully introduced to reveal the mechanisms of TCM, which coincides with the connotation of multi-component, multi-targets, and multi-pathways holistic strategy.16,17
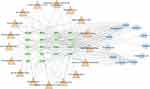
In the present study, network pharmacology, bioinformatics analysis and in vivo experiments were combined to uncover the possible targets and mechanisms of HTD for treating UC, and the detailed flowchart of the study design is shown in Figure 1.
 |
Figure 1 Flowchart of the analysis strategy in the study. |
Materials and Methods
Reagents and Materials
Terra Flava Usta (Batch No: 2006002) was purchased from AnguoJuyaotang Pharmaceutical Co., Ltd. Rehmanniae Radix (Batch No: Y2105018), Scutellariae Radix (Batch No: Y2007053), Aconiti Lateralis Radix Praeparata (Batch No: Y062011002), Asini Corii Colla (Batch No: Y052105035), Atractylodis Macrocephalae Rhizoma (Batch No: Y2006043) and Glycyrrhizae Radix et Rhizoma (Batch No: Y2008055) were purchased from Sichuan Neautus Traditional Chinese Medicine Co., Ltd. HPLC grade methanol was provided by Fisher Scientific (USA). Other chemicals were of analytical grade.
Animals and Diets
The C57BL/6 male mice were provided by the SPF (Beijing) Biotechnology Co., Ltd. (DCXK (Beijing) 2019–0010). The mice (6–8 weeks old, n=24) were randomly divided into 3 groups, and acclimated in independent ventilation cages (IVC) for 6 days with free access to standard mouse chow and tap water in the SPF facility under controlled temperature (22 ± 2°C), humidity (55% ± 10%), and 12 light-dark cycle. Animal experiments were performed according to the Recommended Guidelines for the Care and Use of Laboratory Animals issued by the Chinese Council on Animal Research and approved by the Ethics Committee of Chengdu University of TCM.
Preparation of the HTD Extracts
The HTD extracts were prepared according to the recorded method in “JingGuiYaoLue”. Briefly, the seven herbs Terra Flava Usta (40 g), Rehmanniae Radix (15 g), Scutellariae Radix (15 g), Aconiti Lateralis Radix Praeparata (15 g), Asini Corii Colla (15 g), Atractylodis Macrocephalae Rhizoma (15 g) and Glycyrrhizae Radix et Rhizoma (6 g) were mixed with purified water, and Aconiti Lateralis Radix Praeparata were soaked for 12 hours, while other herbs were soaked for half an hour, then they were boiled for an hour. The filtrate were collected and centrifuged at 3000 rpm for 5 min. The supernatant was harvested and steamed dry, and the residue was dissolved to 5 mL with methanol. The sample was filtered through a 0.22 μm microporous membrane for subsequent testing.
Identification of Active Compounds in HTD
The active compounds of HTD extracts were qualitatively identified by Q Exactive Orbitrap LC-MS/MS (Thermo Fisher Scientific, Waltham, MA, USA). The XBridgeBEHC18 (2.1×100 mm, i.d.; 2.5 μm, Waters, MA, USA) was applied for sample separation at 35°C. Methanol (A) and 0.1% formic acid-water (B) (95:5) were used as mobile phase. Detection wavelength was set at 280 nm. The flow rate and injection volume were set as 0.2 mL/min and 1.0 μL, respectively. Nitrogen was applied as auxiliary gas and sheath gas at a flow rate of 12 L/min. The mass determination was carried out based on positive and negative scanning mode with the m/z of 100–1000.
Acquisition of Corresponding Targets of Active Compounds
All the chemical structures of the identified active compounds were imported into SwissTargetPrediction (http://www.swisstargetprediction.ch/) to predict the targets.18 Targets of these compounds were collected through high-throughput screening and reverse docking.
Acquisition of UC Targets
Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo), a public functional genomics data repository, was used to establish UC targets library.19 Keywords such as “ulcerative colitis” or “ulcer colonitis” were used to search for the known targets related to the pathogenesis of UC from GEO database, so as to find out the difference in gene expression datasets between intestinal tissue samples from patients with UC and normal intestinal tissue samples. Finally, 3 datasets of UC were downloaded from the GEO database: GSE75214 on the (HuGene-1_0-st) Affymetrix Human Gene 1.0 ST Array (GPL6244 platform), including colon tissue samples from 97 UC patients and 11 healthy people; GSE87466 on the (HT_HG-U133_Plus_PM) Affymetrix HT HG-U133+ PM Array Plate (GPL13158 platform), including colon tissue samples from 87 UC patients and 21 healthy people; GSE59071 on the (HuGene-1_0-st) Affymetrix Human Gene 1.0 ST Array (GPL6244 platform), including colon tissue samples from 97 UC patients and 11 healthy people.
The datasets downloaded from the GEO database were preprocessed by background correction, quantile normalization and base logarithm conversions. Finally, the difference in gene expression between normal human and UC patients was clarified. In this study, log FC (fold change) ≥1 and adjusted p-value <0.01 considered that the difference was statistically significant, and the corresponding genes were identified as DEGs, listed in the UC target library.
GO and Pathway Enrichment Analysis
The GO and pathway enrichment analysis was performed through Metascape (https://metascape.org).20 GO functionally annotates key genes into 3 main terms, ie cellular components (CCs), molecular functions (MFs), and biological processes (BPs). Besides, CytoscapeClueGO plugin was used to further analyze the enrichment of BPs. Pathway enrichment analysis unveiled the possible BPs with key targets. In addition, the bubble chart of GO and pathway enrichment analysis were performed on the bioinformatics platform (http://www.bioinformatics.com.cn/).
Construction of Compound-Target-Pathway Network
To characterize the therapeutic mechanisms of HTD for UC, the compound-target-pathway network was constructed using Cytoscape version 3.7.2.21 In the network, the nodes with different colors and shapes represented the drug, compounds, disease, target genes, or disease-related pathways, respectively, and an “edge” was an association between the nodes.
Experimental Validation
Dextran Sodium Sulfate (DSS)-Induced Acute UC Model
Following the previous methods of Mariano et al and Meurer et al22,23 after adaptive feeding for 6 days, the mice in control group (n=8) were fed with water, while those in model group (n=8) and DSS + HTD group (n=8) freely drank 3% DSS (36,000–50,000 Da) solution (prepared by dissolving 3 g of DSS in 100 mL of deionized water). The drinking amount in each group was recorded, and DSS solution was updated every day. Furthermore, the mice in DSS + HTD group were administrated with 2.5 g/kg HTD extracts once a day according to their weight. The drinking amount, body weight, stool properties and blood in stool of mice in each group were recorded every day, and the colitis scores were given by weight loss and stool conditions shown in Table 1. After investigations for one week, the mice in the 3 groups were euthanized, and the serum and colon tissues were collected for further analysis.
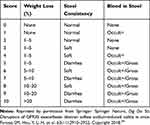 |
Table 1 Scoring Standards of Ulcer Colitis |
Enzyme-Linked Immunosorbent Assay (ELISA)
The levels of inflammatory factors were detected by ELISA kits. The mouse IL-1β ELISA kit (Multi Sciences (Lianke) Biotech, CO., LTD., Hangzhou, China), and mouse IL-6 ELISA kit (Multi Sciences (Lianke) Biotech, CO., LTD., Hangzhou, China) were utilized in accordance with the directions of the manufacture.
Histopathology
The colon samples were cut into small pieces, fixed in 4% paraformaldehyde, embedded in paraffin, and stained with hematoxylin and eosin (H&E). Then histopathological changes were examined under a microscope.
Western Blotting (WB)
Proteins were extracted from the colon tissues using RIPA Lysis Buffer (Wuhan Servicebio Technology Co., Ltd, China) and the protein concentration was determined by bicinchoninic acid (BAC) method on the enzyme labelling instrument (Thermo Fisher Scientific, USA). The SDS-PAGE was used to separate the protein, and subsequently transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were first blocked for 1h with 5% BSA at room temperature, and then probed with primary antibodies of MMP1 (Affinity, dilution of 1:1000), MMP3 (Abcam, dilution of 1:5000), MMP7 (Abcam, dilution of 1:1000), MMP9 (Abcam, dilution of 1:1000), MMP12 (Proteintech, dilution of 1:1000) and β-Actin (Affinity, dilution of 1:5000) at 4°C for overnight and then incubated with HPR-conjugated antibody at 37°C for one hour. Finally, the protein bands were recorded by ELC (Thermo Fish Scientific, USA) and quantified by Image J software to evaluate the protein relative expressions compare with β-actin.
Quantitative Real-Time- PCR (qRT-PCR)
The mRNA expression of UC was detected by qRT-PCR. The colon tissues in each group of mice were pulverized in liquid nitrogen before RNA extraction. The total RNA was isolated and reversely transcribed into cDNA using the RT-qPCR EasyTM (One Step)-SYBR Green I kit (Foregene; Chengdu, China), followed by synthesis via reverse transcription using the Direct RT Mix in the kit, and amplification using Direct qPCR Mix-SYBR of kit according to the manufacturer’s instructions. Then qPCR was carried out on a Bio-Rad real-time PCR system (CFXConnect, USA) for 40 cycles. The expression levels of target genes were normalized to the control housekeeping gene GAPDH. The gene expression was analyzed by the 2−ΔΔCt method. Primer sequences are shown in Table 2.
 |
Table 2 Primer Sequences for qRT-PCR |
Immunohistochemistry and Staining Intensity
Immunolabeling for matrix metalloproteinase-1 (MMP1), MMP3, MMP7, MMP9 and MMP12 was performed on fixed and paraffin-embedded sections. Briefly, following deparaffinization and rehydration, slides were subjected to antigen retrieval in citrate buffer (pH 6.0) for 20 min in a steamer, rinsed in PBS (0.01 mol/L phosphate buffer containing 0.15 mol/L NaCl, pH7.4), and incubated for 10 min in 3% H2O2 to block endogenous peroxidases. After that, the sections were incubated with anti-MMP1 (1:200; Bioss, China), anti-MMP3 (1:200; Bioss, China), anti-MMP7 (1:200; Bioss, China), anti-MMP9 (1:200; Bioss, China), and anti-MMP12 (1:200; Bioss, China) primary antibodies at 4°C overnight in a wet box, and then with HRP-labeled anti-rabbit secondary antibodies for 30 min, followed by counterstaining with hematoxylin, rehydration and mounting. Staining intensity was quantified by Image-Pro Plus 6.0 (Media Cybernetics) and expressed as mean using the formula “mean density = integrated optical density/area of interest”.
Statistical Analysis
Each experiment was repeated 3 times, and the results were expressed as mean ± standard deviation (SD). Significant differences between means were compared using the Bonferroni and Dunnett’s T3 test in the case of homogeneity of variance with the p-value <0.05.
Results
Compounds of HTD Extract and Corresponding Targets
The main compounds of HTD extracts were analyzed qualitatively based on Q Exactive Orbitrap LC-MS/MS. The total ion chromatogram (TIC) is shown under positive-ion polarity mode (A) and negative-ion polarity mode (B) in Figure 2.
 |
Figure 2 Chromatograph of the extracts of HTD by Q Exactive Orbitrap LC-MS/MS on positive-ion polarity mode (A) and negative-ion polarity mode (B). |
More than 60 major compounds were eluted by the Q Exactive Orbitrap LC-MS/MS system from the HTD extracts within 30 min. Among then, a total of 47 compounds were finally identified by comparing with the known chemical constituents in the reported literature data, and deduced according to their mass spectrometry and fragment ion characteristics, as shown in Table 3 and Figure 3.
 |
Table 3 Compositions and Their Product Ions in Huangtu Decoction |
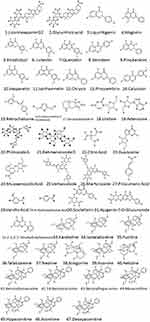 |
Figure 3 Compounds detected in the extracts of HTD by Q Exactive Orbitrap LC-MS/MS. |
A total of 511 corresponding targets of compounds were acquired from Swiss Target Prediction.
Prediction of Targets of HTD for UC
After data preprocessing, a total of 470 DEGs were extracted from 3 datasets (GSE87466, GSE59071, and GSE75214) downloaded from the GEO database, as shown in the volcano map and hot map (Figure 4A and B). Finally, 154 upregulated and 316 downregulated genes were identified in UC tissues compared with normal colon tissues.
Finally, a total of 29 overlapping genes (OGs) of compound targets and UC-related targets, namely HSD17B2, KDR, PLAU, ABCG2, MMP7, NOS2, MMP3, FADS1, CXCR1, AKR1B10, CDC25B, SELL, CD38, MAOA, EDNRA, LRRK2, CA2, ABCB1, STS, PDGFRB, SELP, PTGS2, PFKFB3, PIM2, MMP1, IGFBP5, EPHX2, MMP12 and MMP9, were regarded as potential targets of HTD for the treatment of UC (Figure 4C).
GO and Enrichment Analysis
As shown in Figure 5A and Table 4, the top terms (P<0.01) in the BPs, CCs, and MFs were selected. The enrichment of BPs involved the collagen catabolic process, collagen metabolic process, extracellular matrix disassembly, regulation of neuroinflammatory response, extracellular matrix organization, extracellular structure organization, external encapsulating structure organization, response to oxidative stress, reactive oxygen species metabolic process, and response to inorganic substance. Besides, further analysis of the network was performed by Cytoscape ClueGO plugin, and it was found that the collagen catabolic process, positive regulation of smooth muscle cell proliferation, long-chain fatty acid biosynthetic process, and positive regulation of phospholipase activity were mainly involved in BPs (Figure 5B). The enrichment of CCs mainly involved secretory granule membrane, and other cell components. The enrichment of MFs involved the metalloendopeptidase activity and metallopeptidase activity.
 |
Table 4 GO Functional Enrichment Analysis Results of Overlapping Genes (OGs) |
 |
Figure 5 Bubble diagram of GO functional enrichment analysis of overlapped genes (A) The bubble chart. (B) The visualization analysis of BPs. |
The signal transduction pathways are shown in Table 5 and Figure 6 (P<0.01). The result of pathway enrichment analysis of the HTD for UC revealed that the OGs were significantly associated with MMPs, imatinib, chronic myeloid leukemia, and photodynamic therapy-induced NF-κB survival signaling, in WikiPathways, with IL-4 and IL-13 signaling and collagen degradation in Reactome Gene Sets, and also with MicroRNAs in cancer and IL-17 signaling pathway in KEGG.
 |
Table 5 Pathway Enrichment Analysis Results of Overlapping Genes (OGs) |
 |
Figure 6 Enriched pathway analysis of potential targets of HTD compounds against UC. |
Compound-Target- Pathway Network
As shown in Figure 7, the compound-target-pathway network included 46 nodes (11 compounds, 18 targets, and 17 pathways) and 158 edges. According to the analysis of the network, each active compound acted on at least 2 target genes, each gene was regulated by at least 2 active compounds, and at least 18 genes were potentially involved in each pathway related to UC, indicating the action characteristics of multiple active compounds and targets of HTD in the treatment of UC.
Experimental Validation
Effect of HTD on Histopathological Changes
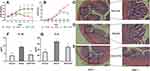
As shown in Figure 8A and B, the mice in model group and DSS+HTD group manifested varying degrees of loose stool and weight loss compared with those in control group. The pathological conditions of colon tissues of the mice treated with DSS+HTD were improved compared with those in model group. The histopathological changes in each group are shown in Figure 8C. The results of H&E staining showed that DSS induced massive inflammatory infiltration, mucosal edema, and hyperplasia in colonic tissues of model group, but less necrosis was observed in DSS+HTD group.
Effect of HTD on Serum Levels of IL-1β and IL-6
As shown in Figure 8F and G, the serum levels of IL-1β and IL-6 were significantly increased in model group compared with those in control group (P<0.01). Compared to the model group, serum IL-1β and IL-6 levels decreased greatly in DSS+HTD group (P<0.01).
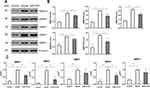
Effect of HTD on MMP Expressions
The expression levels of MMP1, MMP3, MMP7, MMP9 and MMP12 in colon tissues were investigated via immunohistochemical staining (Figure 9) and WB (Figure 10A and B). In contrast with model group, the expressions of MMP1, MMP3 and MMP9 in colon tissues in DSS+HTD group were significantly lower, and the expressions of MMP7 and MMP12 declined with no significant difference. In addition, qRT-PCR was applied to explore the role of HTD in regulating the expression of MMPs in colon tissues. As shown in Figure 10C, DSS significantly upregulated the expressions of MMP1, MMP3, MMP7, MMP9 and MMP12 in colon tissues, but after HTD treatment, the upregulation of MMP1, MMP3 and MMP9 was reversed, while MMP7 and MMP12 had no significant changes.
Discussion
UC is a chronic disease with an increasing incidence worldwide over recent decade, especially in developing nations.2 HTD is a very effective prescription for the clinical treatment of hematochezia in TCM, and has been reported as non-surgical treatment of UC with minimal side effects. However, there is a lack of modern animal experimental evidence and no literature report on its active compounds and mechanism. In this study, the UC model was established to evaluate the therapeutic effects of HTD on UC, and the possible mechanism of this TCM formula for treating UC was also explored based on network pharmacology.
HTD contains 7 medicinal herbs, including Rehmanniae Radix, Scutellariae Radix, Aconiti Lateralis Radix Praeparata, Atractylodis Macrocephalae Rhizoma and Glycyrrhizae Radix et Rhizoma, Terra Flava Usta and Asini Corii Colla. In the present study, HPLC-MS/MS was used to analyze the chemical compositions of HTD, and a total of 47 compounds were identified from it, mostly flavones and alkaloids. Furthermore, network pharmacology was used to explore its possible mechanism. The compound-target-pathway network showed that wogonin with the highest degree was regarded as the most effective compound, followed by LicoricesaponinG2, Liquiritigenin, Retrochalcone, Genistein, Atractylenolide III, Glycyrrhizic acid, Pinobanksin, Pinocembrin, Mussaenosidic acid and Hesperetin. These compounds identified in this formula may provide fundaments for establishing Q-markers for further reliable quality control of HTD. In addition, the possible molecular mechanism and signaling pathway for HTD were also predicted based on GO and KEGG enrichment analysis. The results revealed that the enrichment of BPs of HTD involved collagen catabolic process, collagen metabolic process, and extracellular matrix disassembly. The pathway enrichment analysis results showed that the pathways covered MMPs, MicroRNAs in cancer, IL-17 signaling pathway and so on. Therefore, the prediction results were further verified by in vivo animal experiments, and a similar result to network pharmacology was obtained. HTD was effective in the treatment of UC through decreasing the protein and mRNA expressions of MMP1, MMP3 and MMP9. MMPs are a family of proteolytic enzymes characterized by the ability to degrade extracellular matrix (ECM) proteins such as collagen, proteoglycan, fibronectin, elastin, and laminin.41,42 Chronic inflammation and aberrant tissue remodeling with excessive accumulation or degradation of ECM components are markers in UC. Hence, MMPs are thought to be predominant proteases involved in the pathogenesis of UC.41–43 MMPs have been recognized as noninvasive diagnostic tools for UC in many studies.41,42 Specific synthetic MMP inhibitors and some natural products or therapeutic interventions have been described to ameliorate intestinal inflammation.43
Conclusion
In conclusion, HTD can alleviate the colonic inflammation via inhibiting MMPs including MMP1, MMP3 and MMP9. The present study will benefit the further development of this classic TCM formula.
Acknowledgments
The authors wish to thank the editor and the anonymous reviewers whose constructive comments are very helpful in strengthening the presentation of this paper. The research was supported by Chengdu High-level Key Clinical Specialty Construction Project.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Du L, Ha C. Epidemiology and pathogenesis of ulcerative colitis. Gastroenterol Clin North Am. 2020;49(4):643–654. doi:10.1016/j.gtc.2020.07.005
2. da Silva BC, Lyra AC, Rocha R, et al. Epidemiology, demographic characteristics and prognostic predictors of ulcerative colitis. World J Gastroenterol. 2014;20(28):9458–9467. doi:10.3748/wjg.v20.i28.9458
3. Cohen RD, Yu AP, Wu EQ, et al. Systematic review: the costs of ulcerative colitis in Western countries. Aliment Pharmacol Ther. 2010;31(7):693–707. doi:10.1111/j.1365-2036.2010.04234.x
4. Chen R, Lai LA, Brentnall TA, et al. Biomarkers for colitis-associated colorectal cancer. World J Gastroenterol. 2016;22(35):7882–7891. doi:10.3748/wjg.v22.i35.7882
5. Nehme F, Schneider A, Hamid F. Appendiceal adenocarcinoma associated with ulcerative colitis. ACG Case Rep J. 2019;6(11):e00255. doi:10.14309/crj.0000000000000255
6. Ordás I, Eckmann L, Talamini M, et al. Ulcerative colitis. Lancet. 2012;380(9853):1606–1619. doi:10.1016/s0140-6736(12)60150-0
7. Wehkamp J, Stange EF. Recent advances and emerging therapies in the non-surgical management of ulcerative colitis. F1000 Res. 2018;7:1207. doi:10.12688/f1000research.15159.1
8. Zhang Y, Long Y, Yu S, et al. Natural volatile oils derived from herbal medicines: a promising therapy way for treating depressive disorder. Pharmacol Res. 2021;164:105376. doi:10.1016/j.phrs.2020.105376
9. Xu Q, Guo Q, Wang C, et al. Network differentiation: a computational method of pathogenesis diagnosis in traditional Chinese medicine based on systems science. Artif Intell Med. 2021;118:102134. doi:10.1016/j.artmed.2021.102134
10. Shao MJ, Yan YX, Qi Q, Tang W, Zuo JP. Application of active components from traditional Chinese medicine in treatment of inflammatory bowel disease. Zhongguo Zhong Yao Za Zhi. 2019. 44(3):415–421. doi:10.19540/j.cnki.cjcmm.20180907.001. Chinese. PMID: 30989902.
11. Liu F, Liu Y, Tian C. Effect of Rhizoma Atractylodis extract in protecting gastric mucosa and modulating gastrointestinal immune function in a rat model of spleen deficiency. Nan Fang Yi Ke Da Xue Xue Bao. 2015;35(3):343–7, 354. Chinese. PMID: 25818777.
12. Tang XD, Lu B, Li ZH, et al. Therapeutic effect of Chang’an I recipe (I) on irritable bowel syndrome with diarrhea: a multicenter randomized Double-Blind placebo-controlled clinical trial. Chin J Integr Med. 2018;24(9):645–652. doi:10.1007/s11655-016-2596-9
13. Zhang Q, Duan H, Li R, et al. Inducing apoptosis and suppressing inflammatory reactions in synovial fibroblasts are two important ways for Guizhi-Shaoyao-Zhimu decoction against rheumatoid arthritis. J Inflamm Res. 2021;14:217–236. doi:10.2147/JIR.S287242
14. Manfredi M, Brandi J, Di Carlo C, et al. Mining cancer biology through bioinformatic analysis of proteomic data. Expert Rev Proteomics. 2019;16(9):733–747. doi:10.1080/14789450.2019.1654862
15. Tang S, Jing H, Huang Z, et al. Identification of key candidate genes in neuropathic pain by integrated bioinformatic analysis. J Cell Biochem. 2020;121(2):1635–1648. doi:10.1002/jcb.29398
16. Li ZT, Zhang FX, Fan CL, et al. Discovery of potential Q-marker of traditional Chinese medicine based on plant metabolomics and network pharmacology: periplocae cortex as an example. Phytomedicine. 2021;85:153535. doi:10.1016/j.phymed.2021.153535
17. Pan L, Li Z, Wang Y, et al. Network pharmacology and metabolomics study on the intervention of traditional Chinese medicine Huanglian decoction in rats with type 2 diabetes mellitus. J Ethnopharmacol. 2020;258:112842. doi:10.1016/j.jep.2020.112842
18. Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357–W364. doi:10.1093/nar/gkz382
19. Clough E, Barrett T. The gene expression omnibus database. Methods Mol Biol. 2016;1418:93–110. doi:10.1007/978-1-4939-3578-9_5
20. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019-09234-6
21. Doncheva NT, Morris JH, Gorodkin J, et al. Cytoscape StringApp: network analysis and visualization of proteomics data. J Proteome Res. 2019;18(2):623–632. doi:10.1021/acs.jproteome.8b00702
22. Mariano LNB, Arruda C, Somensi LB, et al. Brazilian green propolis hydroalcoholic extract reduces colon damages caused by dextran sulfate sodium-induced colitis in mice. Inflammopharmacology. 2018;26(5):1283–1292. doi:10.1007/s10787-018-0467-z
23. Meurer MC, Mees M, Mariano LNB, et al. Hydroalcoholic extract of Tagetes erecta L. flowers, rich in the carotenoid lutein, attenuates inflammatory cytokine secretion and improves the oxidative stress in an animal model of ulcerative colitis. Nutr Res. 2019;66:95–106. doi:10.1016/j.nutres.2019.03.005
24. Farooq SM, Hou Y, Li H, et al. Disruption of GPR35 exacerbates dextran sulfate sodium-induced colitis in mice. Dig Dis Sci. 2018;63(11):2910–2922. doi:10.1007/s10620-018-5216-z
25. Song W, Qiao X, Chen K, et al. Biosynthesis-based quantitative analysis of 151 secondary metabolites of licorice to differentiate medicinal Glycyrrhiza species and their hybrids. Anal Chem. 2017;89(5):3146–3153. doi:10.1021/acs.analchem.6b04919
26. Tanaka K, Hayashi K, Fahad A, Arita M. Multi-stage mass spectrometric analysis of saponins in Glycyrrhiza radix. Nat Prod Commun. 2011;6(1):7–10. PMID: 21366035.
27. Chang GH, Bo YY, Cui J, et al. Main chemical constituents in aerial parts of Glycyrrhiza uralensis by UPLC-Q-Exactive Orbitrap-MS. Zhongguo Zhong Yao Za Zhi. 2021;46(6):1449–1459. doi:10.19540/j.cnki.cjcmm.20201225.301
28. Fong SY, Wong YC, Zuo Z. Development of a SPE-LC/MS/MS method for simultaneous quantification of baicalein, wogonin, oroxylin A and their glucuronides baicalin, wogonoside and oroxyloside in rats and its application to brain uptake and plasma pharmacokinetic studies. J Pharm Biomed Anal. 2014;97:9–23. doi:10.1016/j.jpba.2014.03.033
29. Chai CC, Cao Y, Mao M, et al. Comparison of chemical compositions before and after wine-frying of Scutellaria baicalensis based on HPLC characteristic chromatogram, UPLC-Q-TOF/ MS qualitative and multi-component quantitative analysis. Chin Tradit Herb Drugs. 2020;51(9):2436–2447. doi:10.7501/j.issn.0253-2670.2020.09.019
30. Wei Y, Wang SY, Wu SY, et al. Qualitative characterization of flavonoids in Scutellariae Radix by using PREC-IDA-EPI. Zhongguo Zhong Yao Za Zhi. 2018;43(2):345–352. doi:10.19540/j.cnki.cjcmm.20171027.004
31. Cui YY, Zhou YF, Ma YQ, et al. Differences analysis of chemical composition of raw and fried Glycyrrhiza uralensis based on UPLC-QTOF-MS. Chin Pharm. 2020;31(9):1049–1053. doi:10.6039/j.issn.1001-0408.2020.09.06
32. Sun X, Wen HM, Cui XB, et al. Qualitative evaluation of Atractylodis Macrocephalae rhizoma from different habitats by HPLC-PDA fingerprint combined with UFLC-Q-TOF/MS qualitative identification. Chin Tradit Herb Drugs. 2016;47(19):3494–3501. doi:10.7501/j.issn.0253-2670.2016.19.023
33. Cao G, Li Q, Cai H, et al. Investigation of the chemical changes from crude and processed Paeoniae Radix Alba-Atractylodis Macrocephalae rhizoma herbal pair extracts by using Q exactive high-performance benchtop quadrupole-orbitrap LC-MS/MS. Evid Based Complement Alternat Med. 2014;2014:170959. doi:10.1155/2014/170959
34. Zhang BY, Jiang ZZ, Wang YF, et al. Analysis of chemical constituents in fresh, dried and prepared Rehmanniae Radix by UPLC/ESI-Q-TOF MS. Chin Tradit Patent Med. 2016;38(5):1104–1108. doi:10.3969/j.issn.1001-1528.2016.05.029
35. Qiao X, Li R, Song W, et al. A targeted strategy to analyze untargeted mass spectral data: rapid chemical profiling of Scutellaria baicalensis using ultra-high performance liquid chromatography coupled with hybrid quadrupole orbitrap mass spectrometry and key ion filtering. J Chromatogr A. 2016;1441:83–95. doi:10.1016/j.chroma.2016.02.079
36. Wang L, Tan N, Wang H, et al. A systematic analysis of natural alpha-glucosidase inhibitors from flavonoids of Radix scutellariae using ultrafiltration UPLC-TripleTOF-MS/MS and network pharmacology. BMC Complement Med Ther. 2020;20(1):72. doi:10.1186/s12906-020-2871-3
37. Zhang DK, Han X, Rui YL, et al. Analysis on characteristic constituents of crude Aconitum carmichaelii in different regions based on UPLC-Q-TOF-MS. Zhongguo Zhong Yao Za Zhi. 2016;41(3):463–469. doi:10.4268/cjcmm20160318
38. Ye X, Wu J, Zhang D, et al. How Aconiti Radix cocta can treat gouty arthritis based on systematic pharmacology and UPLC-QTOF-MS/MS. Front Pharmacol. 2021;12:618844. doi:10.3389/fphar.2021.618844
39. Zhou SS, Ma ZC, Liang QD, et al. UPLC/Q-TOF-MS-based chemical profiling approach to evaluate the chemical constitution of Radix Aconiti lateralis preparata in the process of decoction. Zhong Xi Yi Jie He Xue Bao. 2012;10(8):894–900. doi:10.3736/jcim20120810
40. Zhu H, Wang C, Qi Y, et al. Fingerprint analysis of Radix Aconiti using ultra-performance liquid chromatography-electrospray ionization/ tandem mass spectrometry (UPLC-ESI/MS n) combined with stoichiometry. Talanta. 2013;103:56–65. doi:10.1016/j.talanta.2012.10.006
41. O’Shea NR, Smith AM. Matrix metalloproteases role in bowel inflammation and inflammatory bowel disease: an up to date review. Inflamm Bowel Dis. 2014;20(12):2379–2393. doi:10.1097/MIB.0000000000000163
42. Maronek M, Marafini I, Gardlik R, et al. Metalloproteinases in inflammatory bowel diseases. J Inflamm Res. 2021;14:1029–1041. doi:10.2147/JIR.S288280
43. de Bruyn M, Vandooren J, Ugarte-Berzal E, et al. The molecular biology of matrix metalloproteinases and tissue inhibitors of metalloproteinases in inflammatory bowel diseases. Crit Rev Biochem Mol Biol. 2016;51(5):295–358. doi:10.1080/10409238.2016.1199535
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.