Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 16
Identification of a tsRNA Contributor to Impaired Diabetic Wound Healing via High Glucose-Induced Endothelial Dysfunction
Authors Zhang XT, Mao ZY, Jin XY, Wang YG, Dong YQ, Zhang C
Received 21 June 2022
Accepted for publication 21 December 2022
Published 31 January 2023 Volume 2023:16 Pages 285—298
DOI https://doi.org/10.2147/DMSO.S379473
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Gian Paolo Fadini
Xiao-Tian Zhang,* Zhen-Yang Mao,* Xiang-Yun Jin, Yu-Gang Wang, Yu-Qi Dong, Chao Zhang
Department of Orthopedics, Renji Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chao Zhang; Yu-Qi Dong, Renji Hospital, Shanghai Jiao Tong University School of Medicine, No. 160, Pujian Road, Pudong New Area, Shanghai, People’s Republic of China, Tel +86-13817307997 ; +86-13331873590, Email [email protected]; [email protected]
Purpose: Delayed skin healing in diabetic wounds is a major clinical problem. The tRNA-derived small RNAs (tsRNAs) were reported to be associated with diabetes. However, the role of tsRNAs in diabetic wound healing is unclear. Our study was designed to explore the tsRNA expression profile and mine key potential tsRNAs and their mechanism in diabetic wounds.
Methods: Skin tissues of patients with diabetic foot ulcers and healthy controls were subjected to small RNA sequencing. The role of candidate tsRNA was explored by loss- and gain-of-function experiments in HUVECs.
Results: A total of 55 differentially expressed tsRNAs were identified, including 12 upregulated and 43 downregulated in the diabetes group compared with the control group. These tsRNAs were mainly concentrated in intercellular interactions and neural function regulation in GO terms and enriched in MAPK, insulin, FoxO, calcium, Ras, ErbB, Wnt, T cell receptor, and cGMP-PKG signaling pathways. tRF-Gly-CCC-039 expression was upregulated in vivo and in vitro in the diabetic model. High glucose disturbed endothelial function in HUVECs, and tRF-Gly-CCC-039 mimics further harmed HUVECs function, characterized by the suppression of proliferation, migration, tube formation, and the expression of Coll1a1, Coll4a2, and MMP9. Conversely, the tRF-Gly-CCC-039 inhibitor could attenuate high-glucose-induced endothelial injury to HUVECs.
Conclusion: We investigated the tsRNAs expression profile in diabetic foot ulcers and defined the impairment role of tRF-Gly-CCC-039 in endothelial function in HUVECs. This study may provide novel insights into accelerating diabetic skin wound healing.
Keywords: diabetic foot ulcers, skin healing, tsRNA expression profile, tRF-28-Q1Q89P9L84DF, endothelial damage
Introduction
Diabetes is an unmet clinical problem with high prevalence that was among predicted to affect 300 million patients by 2025.1 Diabetes is a lifelong metabolic disease, which usually leads to multiple serious complications, including nephropathy, neuropathy, retinopathy, and diabetic non-healing skin ulcers.2 Among them, diabetic non-healing skin ulcers, such as foot ulcers caused by delayed wound healing, are the most expensive and serious complications. Diabetic wounds are the most difficult to cure, which have become a major factor contributing to the health challenge and high cost for the global medical system.3 Therefore, an accurate molecular mechanism analysis of wound healing is essential for improving the therapeutic effect of diabetic wounds.
Angiogenesis, also known as neovascularization or the new blood vessels to nourish damaged tissues, is a vital step in wound healing. Impaired angiogenesis contributes to diminished wound healing in diabetic skin ulcers.4 Therefore, the priority of enhancing wound healing in diabetic skin ulcers is to promote angiogenic responses. Vascular endothelial cells connect the entire vascular system and play a central role in angiogenesis and the wound healing.5 Endothelial dysfunction is also the most basic and earliest pathological hallmark in diabetes.6 However, the current treatment of diabetic skin ulcers is not only expensive but also has side effects, leading to high non-compliance and poor curative effects. Thus, the design of new therapeutic strategies targeting angiogenesis based on vascular endothelial cells may be useful for accelerating diabetic wound healing.
Transfer RNA (tRNA)-derived small RNAs (tsRNAs) are an emerging regulatory class of small noncoding RNA (ncRNA). Transfer halves (tiRNA) and tRNA-derived fragments (tRFs) are the two main categories of tsRNAs,7 which are generated from mature or precursor tRNAs by multiple cleavage of tRNAs.8 tsRNAs can be divided into several types rest with where they map on the precursor or mature tRNA transcript. Based on tRFs’ mapped positions, tRFs are roughly divided into five types: tRF-5, tRF-3, tRF-2, tRF-1, and i-tRF.7 Although the detailed function and mechanism of tsRNAs remain elusive, some paper has shown that tsRNAs play a key role in diabetes and diabetes-related diseases. For example, Yang et al found that altered tsRNAs may play vital roles in the pathogenesis of proliferative diabetic retinopathy.9 Han et al reported the expression profile of tsRNAs using small RNA sequencing (RNA-seq) in the lens epithelium of rats and concluded that tsRNAs may be participated in the pathogenesis of diabetic cataract.10 Monogenic mutations in the TRMT10A gene encoding for a tRNA methyltransferase cause young-onset diabetes. Cosentino et al observed that β-cell death caused by TRMT10A mutations may be attributed to tRF-5.11,12 However, no studies have reported the role of tsRNAs in angiogenesis in diabetic wound healing.
Given the unknown and potential function of angiogenesis and tsRNA mentioned above in diabetic wound healing, this study aimed to profile the tsRNA expression landscape in the skin tissue of diabetic foot ulcers and to uncover the function of key tsRNAs in diabetic wound healing. This study may provide a novel basis for illustrating the mechanism of diabetic wound healing.
Materials and Methods
Samples, Ethic Statement, and Informed Consent
Diabetic patients (n = 5) and healthy control volunteers (n = 5) were recruited from our hospital. The inclusion criteria for patients with diabetes were as follows: age 18–70 years, male or female, extremity trauma developed for more than 1 month without a tendency to heal, and ineffective application of traditional conservative treatment methods, fasting blood glucose ≥7.0 mmol/L, or 2-h postprandial blood glucose ≥11.1 mmol/L. Written informed consent was collected from all patients. The current was approved by the Committee of Ethics at Renji Hospital affiliated to Shanghai Jiaotong University. Non-diabetic patients who did not meet the abovementioned criteria served as the control group. Normal tissues within 5 mm from the trauma edge, without necrotic tissue, were collected. Immediately after isolation, the tissue was placed in lyophilization tubes and stored in liquid nitrogen. The process of isolation to cryopreservation should be less than 1 min.
RNA Isolation
Total RNA was isolated from skin tissue or human umbilical vein endothelial cells (HUVECs) using a TRIzol reagent (Invitrogen Life Technologies, Inc.), according to the manufacturer’s instructions. The integrity and quantity of RNA samples were measured by agarose gel electrophoresis and the Nanodrop™ instrument. For spectrophotometry, the ratio of OD value of A260/A280 and A260/A230 should be close to 2.0 for pure RNA (ratios between 1.8 and 2.1 are acceptable) and more than 1.8, respectively. Quality-qualified RNA (Supplemental Table 1) was stored at −80°C for subsequent experiments.
Small RNA Sequencing and Bioinformatic Analyses
Skin tissue from diabetic foot ulcers (n = 5) and normal skin tissue from healthy control volunteers (n = 5) were subjected to small RNA-seq. In constructing a small RNA-seq library, total RNAs were pretreated by ligating 3′- and 5′-adaptors following m1A and m3C demethylation for efficient reverse transcription. Next, pretreated total RNAs were used for cDNA synthesis and PCR amplification, and then 134–160-bp PCR-amplified fragments were selected. The prepared libraries were qualified and absolutely quantified using Agilent BioAnalyzer 2100. Finally, small RNA-seq was conducted by KangChen Bio-Technology in an Illumina NextSeq 500 system with a 50-bp single end. The RNA-seq data generated in this study are available in the Sequence Read Archive (SRA) under accession number PRJNA893389 on the NCBI.
Raw data of RNA-seq were filtered by FastQC, and trimmed reads (pass Illumina quality filter, trimmed 5′,3′-adaptor bases by cutadapt) were aligned, allowing for one mismatch only to the mature tRNA sequences. Afterward, reads that did not align mature tRNA were mapped to the precursor tRNA sequences, allowing for one mismatch in bowtie.13 The remaining reads were aligned to miRNA reference sequences, allowing for one mismatch in miRDeep2. Sequence annotations of tsRNAs were mapped to the GtRNAdb database (http://gtrnadb.ucsc.edu/), tRFdb database (http://genome.bioch.virginia.edu/trfdb/), and MINIbase database (http://cm.jefferson.edu/MINTbase/). The expression of tsRNAs was calculated using their sequencing counts and normalized as counts per million of total aligned reads. Differentially expressed tsRNAs were screened on the basis of the expression value with R package edgeR. Significance thresholds were |Log2FC| >1.5 and P value <0.05. GO and KEGG were analyzed for differentially expressed tsRNAs.
qRT-PCR Analyses
For tsRNAs, RNAs were prepared using the rtStar™ tRFandtiRNA Pretreatment Kit (Cat# AS-FS-005, Arraystar) and then reverse transcribed into first-strand cDNA and amplified to cDNA using the rtStar™ First-Strand cDNA Synthesis Kit (3’ and 5’ adaptor) (Cat# AS-FS-003, Arraystar), according to the manufacturer’s protocol. For mRNA, reverse transcription into cDNA was conducted by using the RevertAid™ FirstStrand cDNA Synthesis Kit, with DNase I (Cat# K16225 Thermo). The synthesized cDNA was used for RT-qPCR using 2X PCR master mix (Cat# AS-MR-005-5, Arraystar) on the ViiA 7 Real-time PCR System (Applied Biosystems). The 2−ΔΔCq method was used to normalize the gene expression by relative to U6. All primers were synthesized by Sangon Biotech (Shanghai, China) and showed in Supplemental Table 2.
Network Construction
The network of the tsRNA–mRNA pathway was constructed using Cytoscape version 3.6.1. The red diamond, blue oval, and orange oblong indicated tRF-Gly-CCC-039, mRNA, and the pathway, respectively.
Cell Culture and Transfection
HUVECs purchased from Procell (Cat# CL-0122) were cultured in Ham’s F-12K medium (Procell, PM150910) supplemented with 10% fetal bovine serum (10099–141, GIBCO, China), 1% penicillin–streptomycin (E607011, Sangon, China), 0.1 mg/mL of heparin, and 0.03–0.05 mg/mL of endothelial cell growth supplements, at 37°C in a humidified atmosphere with 5% CO2. Twelve hours before the establishment of the diabetic cell model, all mediums were discarded and replaced with phenol red-free low-glucose D-MEM containing 1% calf serum. Then, HUVECs were transferred to Ham’s F-12K medium consisting of either 33 mM high glucose or 5.5 mM normal glucose. For functional analysis, tRF-Gly-CCC-039 mimic/NC and tRF-Gly-CCC-039 inhibitor/NC were transfected into the abovementioned HUVEC-pretreated HG or NG utilizing Lipofectamine 2000 (Invitrogen) agents as per the manufacturer’s protocols.
5-Ethynyl-2′-Deoxyuridine (EdU) Assay
The EdU Cell Proliferation Assay Kit (E607204, Sangon) was used to assess the proliferation of HUVECs. A complete medium of Ham’s F-12K was mixed with EdU solution (1:500) to produce 2× EdU-labeled medium. Next, 3×104/mL HUVECs were seeded in a 24-well plate with 300 μL of 2× EdU-labeled medium at 37°C in a humidified atmosphere with 5% CO2 for 2 h and then washed with phosphate-buffered saline (PBS) to discard residual-free EdU. Cells were fixed with 150 μL of 4% paraformaldehyde at 25°C for 30 min, followed by 150 μL of glycine (2 mg/mL) to neutralize excess paraformaldehyde at 25°C for 5 min. Then, the glycine solution was discarded, followed by treatment with 300 μL of 0.3% Triton X-100 for 10 min at 25°C. Further, cells were coincubated with TAMRA staining solution (prepared in accordance with product specifications) for 30 min at 25°C in a dark environment. Afterward, cells were washed with 300 μL of 0.5% Triton X-100 two times for 10 min at 25°C. Cells were treated with 300 μL of Hoechst solution (5 μg/mL) at 25°C for 20 min in the dark. Finally, cells were rinsed with PBS and the photos were imaged on a fluorescence microscope (OL YMPUS, CKX53).
Wound Healing Assays
The migration ability of HUVECs was evaluated by the wound healing scratch assay. Cells, after transfection, were cultured with a complete medium containing Ham’s F-12K and plated onto a 24-well culture plate, with 1 × 105 cells/well at 37°C in a humidified atmosphere with 5% CO2 for 24 h until a confluent monolayer was formed. Three parallel wells were set. Then, a scratch was made by using a 200-mL pipette tip. After that, the wells were gently washed twice with PBS to discard the individual cells and then replenished with a serum-free culture medium of cytokines. After wounding, the wound-healing images were measured at 0 and 24 h by using a camera (NIKON, Japan).
Tube Formation Assay
The angiogenic activity of HUVECs was assessed by a Matrigel tube-formation assay. One hour before HUVECs were seeded on the 24-well plate, 200 μL of Matrigel (10 mg/mL; Corning, 354234) was added into each well and then placed in an incubator at 37°C for 1 h to solidify. After that, 6 × 104 cells were inoculated on the solidified Matrigel glue and placed in a 5% CO2 cell incubator at 37°C for further cultivation for 18 h. Finally, an inverted phase-contrast microscope (NIKON, Japan) was used to assess tube formation.
Statistical Analyses
Data in this article were exhibited in mean ± standard deviation. GraphPad Prism 7 was used to perform statistical analyses and plots. The t-test was utilized to evaluate the difference between two groups. One-way analysis of variance following Tukey’s test was utilized to evaluate significant differences among four groups. For all data, p less than 0.05 was considered to be statistically significant.
Results
Overview of Small RNA Sequencing
To explore the tsRNAs related to delaying wound healing in patients with diabetes, we performed small RNA-seq on the injured skin tissue of diabetic patients, and healthy tissue served as the control. As a result, yielding a total of >93.3 million Illumina reads, and the Q30 of each sample was above 92%, and the mapped ratios of mature and precursor were low (Supplemental Table 3). These results indicate that the sequencing data were of good quality, which could be used for subsequent analyses.
Identification of tsRNAs
In the present project, a total of 622 tsRNAs were detected, including 152 known tsRNAs from tRFdb (Figure 1A). The Venn diagram showed 272 commonly expressed tsRNAs in two groups, and specifically expressed tsRNAs in the diabetes groups were 52 and 45 in the control groups, respectively (Figure 1B). In addition, tsRNAs were assorted in subtypes based on sites and length. We found that the distribution of subtype tsRNAs was very analogous between the diabetes and control groups, which were primarily enriched in tRF-5c, tRF-1a, tRF-5a, and tRF-3a (Figure 1C). As we known that tRNA isodecoders share the same anticodon, even though their body sequences are different, thereby, from the perspective of tsRNAs, different tRNAs may produce tRFs or tiRNAs with the same sequence, so we showed the number of all subtypes of tsRNAs derived from the same anticodon tRNA in the diabetes and control groups, and the largest number of the tRF-5c subtype was observed in two groups (Figure 1D).
Differential Expression Profiles of tsRNAs Induced by Diabetes
To identify related tsRNAs that involved in the regulation of diabetes, differentially expressed tsRNAs were identified. In total, 55 differentially expressed tsRNAs were recovered, 12 of which were upregulated and 43 were downregulated in the diabetes group compared with the control group (Figure 2A). Among them, the maximum upregulation significance was the greatest for tRF-Gly-CCC-039. Distinguishable tsRNAs expression patterns were identified among the samples by heat map (Figure 2B). To determine the molecular and metabolic functions in which the 57 differentially expressed tsRNAs participate, GO and KEGG analyses were conducted. The differentially expressed tsRNA target-related GO terms include nervous system development, cell migration, axon guidance, cell adhesion, synaptic transmission, homophilic cell adhesion via plasma membrane adhesion molecules, and the neurotrophin TRK receptor signaling pathway (Figure 2C). The GO results indicated that differentially expressed tsRNAs were involved in intercellular interactions and neural function regulation, which are conducive to wound reconstruction and neuropathy. Additionally, the top 20 enriched KEGG pathways showed that differentially expressed tsRNAs targets were mainly enriched in axon guidance, glutamatergic synapse, retrograde endocannabinoid signaling, neurotrophin signaling pathway, MAPK signaling pathway, insulin signaling pathway, FoxO signaling pathway, calcium signaling pathway, Ras signaling pathway, ErbB signaling pathway, Wnt signaling pathway, T cell receptor signaling pathway, and cGMP-PKG signaling pathway (Figure 2D), indicating that differentially expressed tsRNAs are associated with neuropathy, inflammation, and wound renovation.
Network of tRF-Gly-CCC-039
To confirm the reliability of sequencing results, 3 up-regulated and 3 down-regulated differentially expressed tsRNAs were selected for RT-qPCR verification with five independent experiments. Except for tRF-Gly-CCC-018, the expression of the other five differentially expressed tsRNAs was significantly different between the diabetes and control groups, and the results of RT-qPCR analyses were consistent with those of RNA-seq (Figure 3A). Among them, tRF-Gly-CCC-039 and tRF-Phe-GAA-001 were significantly upregulated in the diabetes group, and tRF-Val-CAC-007, tiRNA-Pro-CGG-001, and tiRNA-Val-CAC-003 were significantly downregulated in the diabetes group compared with the control group (Figure 3A). The candidates of tsRNAs were narrowed to tRF-Gly-CCC-039 and tRF-Phe-GAA-001 by focusing on upregulated in the diabetes group. We found that upon in vitro high-glucose stimulation, only tRF-Gly-CCC-039 expression was upregulated compared with the normal glucose group (Figure 3B and C). Therefore, tRF-Gly-CCC-039 was selected for further analysis due to its largest differential multiples in RT-qPCR and RNA-seq results, and it was upregulated both in vivo and in vitro.
Next, a tRF–mRNA–pathway interaction network map was subsequently constructed (Figure 3D). It was predicted that 93 target mRNAs had potential binding sites with tRF-Gly-CCC-039, and of the 93 target mRNAs, 31 were found in KEGG pathway annotation (Figure 3D). Importantly, these targets concentrated typical pathways associated with insulin, inflammation, and diabetic wound healing, such as insulin secretion, insulin signaling pathway, FoxO signaling pathway, TGF-beta signaling pathway, and PI3K-Akt signaling pathway (Figure 3D). These results indicated that tRF-Gly-CCC-039 may be involved in diabetic skin wound healing through the abovementioned target genes and pathways.
tRF-Gly-CCC-039 Supports High-Glucose-Induced Endothelial Injury
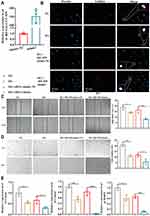
We first blasted tRF-Gly-CCC-039 against MINTbase v2.0, a database for the interactive exploration of nuclear and mitochondrial tRFs (http://cm.jefferson.edu/MINTbase/). In MINTbase v2.0, the molecule of tRF-Gly-CCC-039 hits tRF-28-Q1Q89P9L84DF, which belongs to a class of 28-nt small RNAs, and the sequence is 5′-GCGCCGCTGGTGTAGTGGTATCATGCAA-3′ (Figure 4A). In addition, the expression of tRF-Gly-CCC-039 is enriched in skin cutaneous melanoma (SKCM) and acute myeloid leukemia (Figure 4B), suggesting that tRF-Gly-CCC-039 is a disease-supporting transcript.
To further assess tRF-Gly-CCC-039 function on endothelial dysfunction in diabetes, we constructed a high-glucose-induced endothelial injury model in HUVECs and normal glucose served as the control. Then, the expression of tRF-Gly-CCC-039 was overexpressed by the transfection mimic (Figure 5A). As demonstrated in Figure 5C–E, high-glucose induction resulted in endothelial damage in HUVECs, relative to normal glucose. Importantly, EdU assay unveiled that the tRF-Gly-CCC-039 mimic significantly suppressed the proliferation of HUVECs under high-glucose conditions (Figure 5B). The role of tRF-Gly-CCC-039 in endothelial cell migration was determined by using wound-healing assay, and the results showed that under high-glucose conditions, the tRF-Gly-CCC-039 mimic significantly inhibited the motility of HUVECs (Figure 5C). To investigate the role of tRF-Gly-CCC-039 in the angiogenic capacity of HUVECs, we performed the tube formation assays. The results revealed a significant reduction in the number of tubes in the tRF-Gly-CCC-039 mimic groups compared with the NC group under high-glucose conditions (Figure 5D). At the molecular level, compared with normal glucose groups, matrix-associated gene expression, such as Col1a1, Col4a2, and MMP9, declined after high-glucose induction, and a further decline was observed after tRF-Gly-CCC-039 mimic treatment (Figure 5E). Therefore, these results suggested that tRF-Gly-CCC-039 mimic transfection aggravated high-glucose-induced impairment in HUVECs.
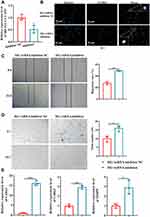
In addition, the damage to tRF-Gly-CCC-039 in endothelial cells induced by high glucose was supported by in vitro inhibition experiments. The tRF-Gly-CCC-039 inhibitor significantly decreased tRF-Gly-CCC-039 expression in HUVECs (Figure 6A). As shown in Figure 6B–E, proliferation, migration, tube formation, and matrix-associated gene expression (Col1a1, Col4a2, and MMP9) were significantly elevated under tRF-Gly-CCC-039 inhibitor treatment rather than inhibitor NC in high-glucose-induced HUVECs. Taken together, tRF-Gly-CCC-039 is a permissive transcript for endothelial injury induced by high glucose.
Discussion
Diabetes is a multifaceted metabolic disease, and diabetes are frequently present with poor wound healing.14 Current therapeutic routes are limited, underlining the need for an in-depth understanding of the pathological mechanism and new therapeutic targets. In this study, we characterized the tsRNAs expression profile in the skin tissue of diabetic foot ulcers and healthy controls and uncovered 57 differentially expressed tsRNAs, which may be participated in wound repair. We also proved that tRF-Gly-CCC-039 mimic transfection aggravated high-glucose-induced impairment in HUVECs, manifested by the reduced proliferation, migration, tube formation, and expression of Col1a1, Col4a2, and MMP9, whereas treatment with the tRF-Gly-CCC-039 inhibitor the opposite effect. Taken together, 57 altered tsRNAs may be involved in wound healing among people with diabetes, and tRF-Gly-CCC-039 inhibits diabetic wound healing through angiogenesis. To our knowledge, this is the second report on the tsRNA profile in diabetic wound healing and repressive effects of tRF-Gly-CCC-039 on diabetic wound healing were also first revealed. Our data have provided novel insights into the tRF-Gly-CCC-039-modulating role of angiogenesis in diabetic wound healing.
In the present study, we found that overexpression of tRF-Gly-CCC-039 HUVECs slowed proliferation and migration, whereas the inhibition of tRF-Gly-CCC-039 showed the opposite result. tRF-Gly-CCC-039, also known as tRF-28-Q1Q89P9L84DF, originated from the 5′-end of tRNA-Gly-CCC. tRNA-Gly genes have three distinct but related families, including -CCC, -GCC, and -TCC anticodons, which are conserved in many mammals.15 Among them, the tRFs1 derived from tRNA-Gly-GCC has been demonstrated to promote cardiomyocyte hypertrophy by targeting Timp3 expression and intergenerational inheritance.16 piR-61648 (tRFs-like, maybe derive from the 5′-arm of tRNA-Gly-GCC-2) is the richest piRNA in human saliva,17 and it is upregulated in patients with human hepatitis B,18 and its somatic tissue specificity may play a role in translational control.19 These results implicated that the tissue-specific expression of tRFs derived from the 5′-tRNA-Gly-GCC is closely related to various diseases. However, very few studies have reported the function of tRF-Gly-CCC. Sharma et al found that the abundance of 5′-fragments of tRNA-Gly, including tRF-Gly-CCC, tRF-Gly-GCC, and tRF-Gly-TCC, exhibited a significant increase in low-protein diet sperm compared with control sperm, and upon a low-protein diet induction, tRF-Gly-GCC can regulate endogenous retroelement MERVL-driven transcript expression in early embryos.20 Based on the highly conservative tRF and previous research results, we hypothesized that the upregulation of tRF-Gly-CCC-039 in diabetic skin tissue is related to the slow healing of diabetic wounds. Our cell experiment results supported this hypothesis. Similarly, Yan et al reported that 22 tsRNAs were differentially expressed among normal skin tissue, diabetic foot ulcers, and hyperbaric oxygen-treated skin tissue.21 Moreover, tsRNAs have been proven to be involved in human hypertrophic scar formation.22 In summary, tRF-Gly-CCC-039 is a permissive transcript for endothelial injury in damaged diabetic skin tissue. Unfortunately, no more studies have been conducted on the role of tsRNA in wound healing or diabetes. We will do more to that end in the future.
In the present study, KEGG analysis indicated that tRF-Gly-CCC-039 and the differentially expressed tsRNAs were related to the classical signaling pathway of insulin, inflammation, and diabetic wound healing, such as insulin secretion, insulin, FoxO, TGF-beta, MAPK, and PI3K-Akt signaling pathway. It is worth mentioning that these pathways are linked to diabetic wound healing. For example, the significance of insulin secretion and insulin signaling pathway in diabetes is proven beyond doubt. Additionally, the MAPK and PI3K-Akt pathways are part of the insulin signaling cascade.23,24 The FoxO and TGF-beta signaling pathways have also been shown to be involved in diabetic wound healing. Huang et al demonstrated that resveratrol can promote angiogenesis to improve diabetic wound healing by inhibiting FOXO1-c-Myc expression.25 The TGF-beta family is important in inflammation, angiogenesis, reepithelialization, and connective tissue regeneration. Among them, TGF-beta3 promotes keratinocyte migration and recruits inflammatory cells and fibroblasts to the wound site, which facilitates wound healing.26 Moreover, interactions may be observed among these molecular signals that promote wound healing, for instance, insulin and TOR signal in parallel with FoxO and S6K promote epithelial wound healing. In summary, these signaling pathways involved in the target genes of tRF-Gly-CCC-039, such as insulin secretion, insulin, FoxO, TGF-beta, MAPK, PI3K-Akt signaling pathway, etc., are significant pathways for diabetic wound healing, indicating that tRF-Gly-CCC-039 plays a vital role in diabetic wound healing.
To mimic angiogenesis in vitro, we chose to use HUVECs. HUVEC is a classic and widely used cell model system to study angiogenesis.27,28 The advantage of the HUVEC model is manifested in four main aspects. First, HUVECs are relatively and easily accessible endothelial cell types that can be isolated in a standard manner or purchased commercially. Second, HUVECs express numerous important vascular endothelial cell markers, such as CD106, ICAM-1, VEFGR2, VE-cadherin, and NOS.29–31 Third, HUVECs have been shown to respond to a variety of physiological and/or pathological stimuli, such as high glucose,32 hypoxia,33 and shear stress.34 However, despite these advantages, the differences between HUVECs and endothelial cells participating in angiogenesis should be noted. For example, HUVECs do not respond to palmitic acid stimulation, which differs from human microvascular endothelial cells.35,36 Fortunately, the study confirmed that the high-glucose-induced phenotypic changes in HUVECs are also applicable to mice.25,28 Therefore, the applicability of the HUVEC model for angiogenesis studies can be recognized but should be vigilantly validated for different stimuli.
Conclusion
In conclusion, we analyzed global tsRNAs expression levels and obtained 57 differentially expressed tsRNAs in damaged skin tissue of patients with diabetes by using small RNA-seq. GO and KEGG pathway analyses revealed that these differentially expressed tsRNAs were involved in many pathophysiological processes, including neuropathy, insulin secretion, wound renovation, and inflammatory responses. Moreover, tRF-Gly-CCC-039 exerted an inhibitory effect on HUVEC behavior. Our study has widespread implications, and it provides abundant tsRNA resources for wound healing in people with diabetes.
Data Sharing Statement
All the data can be found in the main paper or additional supporting files.
Ethics Approval and Informed Consent
The study was reviewed and approved by the Institutional Review Board at Renji Hospital affiliated to Shanghai Jiaotong University. Written informed consents were collected from all patients.
Consent for Publication
All authors confirmed consent for publication.
Funding
There is no funding to report.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Larijani B, Ghahari A, Warnock GL, et al. Human fetal skin fibroblasts: extremely potent and allogenic candidates for treatment of diabetic wounds. Med Hypotheses. 2015;84(6):577–579. doi:10.1016/j.mehy.2015.03.004
2. Zheng Y, Ley SH, Hu FB. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat Rev Endocrinol. 2018;14(2):88–98. doi:10.1038/nrendo.2017.151
3. Frykberg RG, Banks J. Challenges in the treatment of chronic wounds. Adv Wound Care. 2015;4(9):560–582. doi:10.1089/wound.2015.0635
4. Ackermann M, Pabst AM, Houdek JP, et al. Priming with proangiogenic growth factors and endothelial progenitor cells improves revascularization in linear diabetic wounds. Int J Mol Med. 2014;33(4):833–839. doi:10.3892/ijmm.2014.1630
5. Kruger-Genge A, Blocki A, Franke RP, et al. Vascular endothelial cell biology: an update. Int J Mol Sci. 2019;20(18):4411. doi:10.3390/ijms20184411
6. Jiang M, Huang S, Duan W, et al. Inhibition of acid sphingomyelinase activity ameliorates endothelial dysfunction in db/db mice. Biosci Rep. 2019;39(4). doi:10.1042/BSR20182144.
7. Wen JT, Huang ZH, Li QH, et al. Research progress on the tsRNA classification, function, and application in gynecological malignant tumors. Cell Death Discov. 2021;7(1):388. doi:10.1038/s41420-021-00789-2
8. Liu Q, Ding C, Lang X, et al. Small noncoding RNA discovery and profiling with sRNAtools based on high-throughput sequencing. Brief Bioinform. 2021;22(1):463–473. doi:10.1093/bib/bbz151
9. Yang Y, Yue W, Wang N, et al. Altered expressions of transfer RNA-derived small RNAs and microRNAs in the vitreous humor of proliferative diabetic retinopathy. Front Endocrinol. 2022;13:913370. doi:10.3389/fendo.2022.913370
10. Han X, Cai L, Lu Y, et al. Identification of tRNA-derived fragments and their potential roles in diabetic cataract rats. Epigenomics. 2020;12(16):1405–1418.
11. Cosentino C, Toivonen S, Diaz Villamil E, et al. Pancreatic beta-cell tRNA hypomethylation and fragmentation link TRMT10A deficiency with diabetes. Nucleic Acids Res. 2018;46(19):10302–10318. doi:10.1093/nar/gky839
12. Jacovetti C, Bayazit MB, Regazzi R. Emerging classes of small non-coding RNAs with potential implications in diabetes and associated metabolic disorders. Front Endocrinol. 2021;12:670719. doi:10.3389/fendo.2021.670719
13. Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi:10.1186/gb-2009-10-3-r25
14. Fan Y, Wu W, Lei Y, et al. Edaravone-loaded alginate-based nanocomposite hydrogel accelerated chronic wound healing in diabetic mice. Mar Drugs. 2019;17(5):285. doi:10.3390/md17050285
15. Tang DT, Glazov EA, McWilliam SM, et al. Analysis of the complement and molecular evolution of tRNA genes in cow. BMC Genomics. 2009;10:188. doi:10.1186/1471-2164-10-188
16. Shen L, Gan M, Tan Z, et al. A novel class of tRNA-derived small non-coding RNAs respond to myocardial hypertrophy and contribute to intergenerational inheritance. Biomolecules. 2018;8(3):54. doi:10.3390/biom8030054
17. Ogawa Y, Taketomi Y, Murakami M, et al. Small RNA transcriptomes of two types of exosomes in human whole saliva determined by next generation sequencing. Biol Pharm Bull. 2013;36(1):66–75. doi:10.1248/bpb.b12-00607
18. Zhang Y, Zhang Y, Shi J, et al. Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection. J Mol Cell Biol. 2014;6(2):172–174. doi:10.1093/jmcb/mjt052
19. Torres AG, Reina O, Stephan-Otto Attolini C, et al. Differential expression of human tRNA genes drives the abundance of tRNA-derived fragments. Proc Natl Acad Sci U S A. 2019;116(17):8451–8456. doi:10.1073/pnas.1821120116
20. Sharma U, Conine CC, Shea JM, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351(6271):391–396. doi:10.1126/science.aad6780
21. Yan Z, Cui X, Huang M, et al. Integrated analysis of tRNA-derived small RNAs reveals new therapeutic genes of hyperbaric oxygen in diabetic foot ulcers. Epigenomics. 2021;13(22):1817–1829. doi:10.2217/epi-2021-0284
22. Zhang Y, Deng Q, Tu L, et al. tRNAderived small RNAs: a novel class of small RNAs in human hypertrophic scar fibroblasts. Int J Mol Med. 2020;45(1):115–130. doi:10.3892/ijmm.2019.4411
23. Li T, Mo H, Chen W, et al. Role of the PI3K-akt signaling pathway in the pathogenesis of polycystic ovary syndrome. Reprod Sci. 2017;24(5):646–655. doi:10.1177/1933719116667606
24. Wu C, Jiang F, Wei K, et al. Exercise activates the PI3K-AKT signal pathway by decreasing the expression of 5alpha-reductase type 1 in PCOS rats. Sci Rep. 2018;8(1):7982. doi:10.1038/s41598-018-26210-0
25. Huang X, Sun J, Chen G, et al. Resveratrol promotes diabetic wound healing via SIRT1-FOXO1-c-Myc signaling pathway-mediated angiogenesis. Front Pharmacol. 2019;10:421. doi:10.3389/fphar.2019.00421
26. Barrientos S, Stojadinovic O, Golinko MS, et al. Growth factors and cytokines in wound healing. Wound Repair Regen. 2008;16(5):585–601.
27. Cao Y, Gong Y, Liu L, et al. The use of human umbilical vein endothelial cells (HUVECs) as an in vitro model to assess the toxicity of nanoparticles to endothelium: a review. J Appl Toxicol. 2017;37(12):1359–1369. doi:10.1002/jat.3470
28. Bi H, Li H, Zhang C, et al. Stromal vascular fraction promotes migration of fibroblasts and angiogenesis through regulation of extracellular matrix in the skin wound healing process. Stem Cell Res Ther. 2019;10(1):302. doi:10.1186/s13287-019-1415-6
29. Li H, Cai E, Cheng H, et al. FGA controls VEGFA secretion to promote angiogenesis by activating the VEGFR2-FAK signalling pathway. Front Endocrinol. 2022;13:791860. doi:10.3389/fendo.2022.791860
30. Huang MZ, Yang YJ, Liu XW, et al. Aspirin eugenol ester attenuates oxidative injury of vascular endothelial cells by regulating NOS and Nrf2 signalling pathways. Br J Pharmacol. 2019;176(7):906–918. doi:10.1111/bph.14592
31. Chen T, Zhang X, Zhu G, et al. Quercetin inhibits TNF-alpha induced HUVECs apoptosis and inflammation via downregulating NF-kB and AP-1 signaling pathway in vitro. Medicine. 2020;99(38):e22241. doi:10.1097/MD.0000000000022241
32. Lin F, Yang Y, Wei S, et al. Hydrogen sulfide protects against high glucose-induced human umbilical vein Endothelial cell injury through activating PI3K/Akt/eNOS pathway. Drug Des Devel Ther. 2020;14::621–633. doi:10.2147/DDDT.S242521
33. Chai M, Gu C, Shen Q, et al. Hypoxia alleviates dexamethasone-induced inhibition of angiogenesis in cocultures of HUVECs and rBMSCs via HIF-1alpha. Stem Cell Res Ther. 2020;11(1):343. doi:10.1186/s13287-020-01853-x
34. Dong G, Yang S, Cao X, et al. Low shear stress-induced autophagy alleviates cell apoptosis in HUVECs. Mol Med Rep. 2017;15(5):3076–3082. doi:10.3892/mmr.2017.6401
35. Maloney E, Sweet IR, Hockenbery DM, et al. Activation of NF-kappaB by palmitate in endothelial cells: a key role for NADPH oxidase-derived superoxide in response to TLR4 activation. Arterioscler Thromb Vasc Biol. 2009;29(9):1370–1375. doi:10.1161/ATVBAHA.109.188813
36. Cheng AM, Handa P, Tateya S, et al. Apolipoprotein A-I attenuates palmitate-mediated NF-kappaB activation by reducing Toll-like receptor-4 recruitment into lipid rafts. PLoS One. 2012;7(3):e33917. doi:10.1371/journal.pone.0033917
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.