Back to Journals » OncoTargets and Therapy » Volume 14
Identification of a Novel SLC8A1-ALK Fusion and Non-Canonical Expression Significantly Responding to ALK-TKIs in Lung Adenocarcinoma: A Case Report
Authors Zhu X, He Y, Wang Y , Lei Y, Su X, Liu Y , Wu S , He Z
Received 20 June 2021
Accepted for publication 9 September 2021
Published 27 September 2021 Volume 2021:14 Pages 4915—4920
DOI https://doi.org/10.2147/OTT.S319845
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Xingyu Zhu,1,* Yuqi He,2,* Yin Wang,3,* Yan Lei,3 Xiaoxing Su,3 Yifan Liu,1 Shuangxiu Wu,3 Zhengfu He1
1Department of Thoracic Surgery, Sir Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, 310006, People’s Republic of China; 2Monash School of Medicine, Monash University, Clayton, VIC, 3800, Australia; 3Berry Oncology Corporation, Beijing, 102206, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Shuangxiu Wu
Berry Oncology Corporation, Beijing, 102206, People’s Republic of China
Email [email protected]
Zhengfu He
Department of Thoracic Surgery, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, 3 East Qing Chun Road, Hangzhou, 310006, Zhejiang, People’s Republic of China
Tel/Fax +86 571-86006156
Email [email protected]
Background: Approximately 2– 7% of patients with non-small cell lung cancer harbor anaplastic lymphoma kinase (ALK) rearrangement events. Of note, typical ALK actionable rearrangements are sensitive to treatment with tyrosine kinase inhibitors (TKIs). However, different types of ALK fusion influence the clinical outcomes of this therapeutic approach. Approximately 10– 40% of patients with ALK-fusion positive non-small cell lung cancer do not response to ALK-TKI therapy. Therefore, it is important to accurately identify the types of ALK rearrangement for appropriate selection of clinical treatment.
Case Report: Using a DNA-targeted next-generation sequencing technique, we found a novel solute carrier family 8 member A1 (SLC8A1)-ALK fusion type in a patient with lung adenocarcinoma. Further reverse transcriptase-polymerase chain reaction and Sanger sequencing demonstrated the rearrangement as a B-cell CLL/lymphoma 11A (BCL11A)-ALK fusion at the transcriptional level. The patient showed a rapid and strong response to treatment with crizotinib, which lasted for 9 months. The patient also responded well to treatment with alectinib after developed resistance to crizotinib.
Conclusion: A strategy combining DNA-targeted next-generation sequencing with RNA reverse transcriptase-polymerase chain reaction and sequencing, besides fluorescence in situ hybridization and immunohistochemistry, may provide an effective and practical solution for correct identification of partner genes and fusion structures in the diagnosis of ALK rearrangements, particularly for non-canonical expression patterns of ALK fusion events. The combined approach may lead to more benefits for patients.
Keywords: lung adenocarcinoma, anaplastic lymphoma kinase rearrangement, tyrosine kinase inhibitor, treatment, case
Introduction
In non-small cell lung cancer (NSCLC), approximately 2–7% of patients harbor anaplastic lymphoma kinase (ALK) gene rearrangement events. These events often result in aberrant expression and oncogenic activation of ALK driven by the promotion of fusion-partner genes.1,2 With the development of targeted therapy using tyrosine kinase inhibitors (TKIs), ALK-TKIs (eg, alectinib and crizotinib) have markedly improved the clinical outcomes of patients with advanced ALK fusion-positive NSCLC.3–7 Thus far, >90 ALK rearrangement types have been found.8 In addition, approximately half of ALK fusions are non-canonical or complex, which have been associated with poor clinical response to ALK-TKI therapy, accounting for 10–40% of ALK rearrangement events.9 Fusions that upregulate the expression of the entire ALK domain, such as rearrangements with EMAP like 4 (EML4), nucleophosmin (NPM), tropomyosin 3 (TPM3), trafficking from ER to golgi regulator (TFG), 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (ATIC), clathrin heavy chain (CLTC), etc, have been linked to good responses to ALK-TKI therapy.3–5,8,9 Thus, the clinical outcomes of therapy with ALK-TKIs vary depending on the type of fusion. Therefore, it is necessary to accurately detect ALK rearrangement events for proper selection of clinical treatment. In this article, we report a rare solute carrier family 8 member A1 (SLC8A1)-ALK fusion type identified using DNA targeted next-generation sequencing (NGS) and further examined the expression of the entire ALK domain on RNA levels in a patient with lung adenocarcinoma; we also report on the follow-up of treatment with ALK-TKIs.
Case Presentation
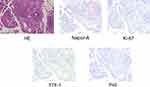
A 62-year-old Chinese man without smoking history was admitted to Sir Run Run Shaw Hospital in September 2019 because of cough with phlegm lasting for 2 months and presence of blood in the phlegm. On physical examination, there was no dysmorphic features. Chest enhanced computed tomography (CT) scan showed a mass at the right lower lobe, multiple nodular thickening with right horizontal fissures, and local atelectasis in the middle and lower lobes of the right lung, accompanied by bilateral pleural effusion and inflammatory lesions (more severe on the right side) and plump mediastinal lymph-nodes. Further examination did not detect metastatic lesions. Subsequently, CT-guided biopsy of the lung tumor tissue was performed. Pathological examination showed NSCLC, and the clinical analysis indicated stage IVA (cT3N2M1a). The immunohistochemical analysis revealed positive expression for napsin A, Ki-67, thyroid transcription factor 1 (TTF-1), and negative expression for P40 (Figure 1), indicating an adenocarcinoma origin of the lung tumor.
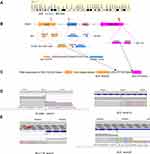
To determine the possible presence of somatic mutations suitable for targeted therapy, DNA-level NGS was also performed on the biopsy specimen based on a 10-gene panel (including ALK, BRAF, EGFR, ERBB2, KRAS, MET, PIK3CA, RET, ROS1 and TP53, Berry Oncology Corporation, Beijing, China). A novel solute carrier family 8 member A1 (SLC8A1)-ALK fusion type was identified with a mutant allele frequency of 5.98%. The sequencing results showed that the SLC8A1 exon 2 rearranged with ALK exon 19 (Figure 2A, B and D). The breakpoints and corresponding fusion sequences were further examined using reverse transcriptase-polymerase chain reaction (RT-PCR), followed by Sanger sequencing to confirm its expression pattern on RNA levels. Of note, the results of targeted RT-PCR sequencing revealed a different ALK rearrangement partner gene: a B-cell CLL/lymphoma 11A (BCL11A) exon 3 rearranged with ALK exon 20 (Figure 2A, C and E). The transcript fusion type retained the N-terminal part of BCL11A and the entire kinase domain of ALK, which may lead to transformation of constitutive kinase activity and carcinogenesis.
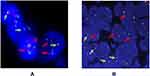
Fluorescence in situ hybridization was performed to confirm the ALK rearrangement events on a chromosomal level in the biopsy specimen obtained from the patient (Figure 3A), with typical positive ALK-rearrangement signals in comparison with the positive reference EML4-ALK fusion image (Figure 3B). Apart from the aforementioned ALK fusion, there was no epidermal growth factor receptor (EGFR), TP53 and KRAS mutations or ROS1 rearrangement detected in this patient.
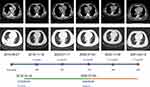
The patient was treated with crizotinib (dose: 250 mg, twice daily) from October 16, 2019. Chest CT scan images demonstrated that the mass, plump mediastinal lymph nodes and pleural effusion had obviously resolved at 1 month and reached stable disease status at 3 months after the treatment (Figure 4). However, a chest CT scan (July 4, 2020) revealed an increased solid composition of the mass, multiple thickened nodules with right horizontal fissures, and plum per mediastinal lymph-nodes appeared in comparison with previous CT scan images (January 11, 2020) (Figure 4). We considered that the patient developed progressive disease during the treatment with crizotinib, and the progression-free survival was 9 months. Thenceforward, alectinib was administered (dose: 600 mg, twice daily) from July 4, 2020. A chest CT scan performed after 4 and 8 months showed partial response. The patient remains under surveillance up to now.
Discussion
To the best of our knowledge, this is the first reported case of the SLC8A1 exon 2-ALK exon 19 fusion type. Fluorescence in situ hybridization analysis also confirmed the positive ALK-rearrangement signals on DNA levels. However, the response of SLC8A1-ALK fusion to treatment with ALK-TKIs remains unknown. The response of TKI treatments for ALK-rearrangement patients largely depends on whether the ALK domain is expressed entirely and at high levels driven by the rearranged-partner genes, which are influenced by complex rearrangement structures and RNA splicing mechanisms, and sometimes disaccord on DNA levels and RNA levels.9 Therefore, RT-PCR and Sanger sequencing were performed to validate the transcription pattern of the fusion gene and confirm the sequences around the breaking point. Notably, the RT-PCR sequencing demonstrated a different type of ALK fusion transcript (ie BCL11A exon 3-ALK exon 20 fusion) in this patient (Figure 3). Fortunately, this ALK-fusion expression pattern is similar to that of BCL11A exon 4-ALK exon 20, which was recently reported to respond strongly to the treatment with crizotinib in NSCLC.10 This finding suggested that ALK-TKI treatment may theoretically benefit the patient of this case. Thus, we administered him crizotinib, and the response was prompt and markedly in the first 9 months (Figure 4). Following the development of resistance to crizotinib, treatment with alectinib was initiated. Thus far, this therapy remains effective, suggesting that alectinib may be an alternative option in patients with this ALK-fusion type after development of resistance to crizotinib.
The exact mechanism through which the ALK fusion participates in carcinogenesis remains to be determined. Nevertheless, it is accepted that the fusion gene can upregulate the expression of ALK and increase its activity, thereby activating ALK self-phosphorylation and further triggering downstream oncogenic signaling pathways.1,2 As reported by an increasing number of studies, the aberrant kinase activity varies depending on the structures of different ALK rearrangements.9 Different fusion structures influence the integrity of the kinase domain, intracellular localization and stability of the fusion protein.11–13 Moreover, they may lead to fusion with different partner genes at the transcriptional levels and DNA levels mainly due to complex RNA splicing mechanisms during expression, as shown in this case. These influencing factors resulted in variable clinical responses to treatment with ALK-TKIs ranging from sensitivity to resistance.9,14–16 Notably, some coexistent gene mutations, such as TP53 mutations, also impair the efficacy of ALK-TKI treatment.9
Conclusion
Using a targeted DNA sequencing panel, we report a NSCLC patient harboring a novel SLC8A1-ALK fusion. Further RT-PCR and Sanger sequencing demonstrated this ALK-fusion type expressed as another similarly established BCL11A-ALK fusion pattern at the transcriptional levels. The results assisted in the drug selection of ALK-TKI treatment, which eventually benefited the patient. Therefore, a strategy combining DNA-based NGS with RNA-based RT-PCR sequencing should be considered in clinical practice for the accurate diagnosis of ALK rearrangements.
Abbreviations
NSCLC, non-small cell lung cancer; ALK, anaplastic lymphoma kinase; TKIs, tyrosine kinase inhibitors; NPM, nucleophosmin; TPM3, tropomyosin 3; TFG, trafficking from ER to golgi regulator; ATIC, 5- aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase; CLTC, clathrin heavy chain; SLC8A1, solute carrier family 8 member A1; NGS, targeted next-generation sequencing; CT, computed tomography; RT-PCR, reverse transcriptase-polymerase chain reaction; TTF-1, thyroid transcription factor 1; BCL11A, B-cell CLL/lymphoma 11A; EGFR, epidermal growth factor receptor.
Data Sharing Statement
The data and materials in this case are available from the corresponding author on reasonable request.
Ethics
Written informed consent was obtained from the patient for publication of this case report and any accompanying images. This study protocol was reviewed and the need for approval was waived by Ethics Committee of Sir Run Run Shaw Hospital.
Funding
The work was supported by the Major Research Project of Science Technology, Department of Zhejiang Province (grant number: 2021C03124); Wu Jieping Medical foundation (320.6750.19092-12).
Disclosure
Yin Wang, Xiaoxing Su, Yan Lei and Shuangxiu Wu are employees of Berry Oncology Corporation. The authors report no other conflicts of interest in this work.
References
1. Solomon B, Varella-Garcia M, Camidge DR. ALK gene rearrangements: a new therapeutic target in a molecularly defined subset of non-small cell lung cancer. J Thorac Oncol. 2009;4(12):1450–1454. doi:10.1097/JTO.0b013e3181c4dedb
2. Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566. doi:10.1038/nature05945
3. Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014;371(23):2167–2177. doi:10.1056/NEJMoa1408440
4. Wu YL, Lu S, Lu Y, et al. Results of PROFILE 1029, a Phase III comparison of first-line crizotinib versus chemotherapy in East Asian patients with ALK-positive advanced non-small cell lung cancer. J Thorac Oncol. 2018;13(10):1539–1548. doi:10.1016/j.jtho.2018.06.012
5. Zhang YC, Zhou Q, Wu YL. Efficacy of crizotinib in first-line treatment of adults with ALK-positive advanced NSCLC. Expert Opin Pharmacother. 2016;17(12):1693–1701. doi:10.1080/14656566.2016.1208171
6. Shaw AT, Gandhi L, Gadgeel S, et al. Alectinib in ALK-positive, crizotinib-resistant, non-small-cell lung cancer: a single-group, multicentre, Phase 2 trial. Lancet Oncol. 2016;17(2):234–242. doi:10.1016/S1470-2045(15)00488-X
7. Hida T, Nokihara H, Kondo M, et al. Alectinib versus crizotinib in patients with ALK-positive non-small-cell lung cancer (J-ALEX): an open-label, randomised phase 3 trial. Lancet. 2017;390(10089):29–39. doi:10.1016/S0140-6736(17)30565-2
8. Ou SHI, Zhu VW, Nagasaka M. Catalog of 5ʹ fusion partners in ALK+ NSCLC circa 2020. JTO Clin Res Rep. 2020;1(1):100015. doi:10.1016/j.jtocrr.2020.100015
9. Kang J, Zhang XC, Chen HJ, et al. Complex ALK fusions are associated with better prognosis in advanced non-small cell lung cancer. Front Oncol. 2020;10(2793):596937. doi:10.3389/fonc.2020.596937
10. Tian Q, Deng WJ, Li ZW. Identification of a novel crizotinib-sensitive BCL11A-ALK gene fusion in a nonsmall cell lung cancer patient. Eur Respir J. 2017;49(4):1602149. doi:10.1183/13993003.02149-2016
11. Bayliss R, Choi J, Fennell DA, Fry AM, Richards MW. Molecular mechanisms that underpin EML4-ALK driven cancers and their response to targeted drugs. Cell Mol Life Sci. 2016;73(6):1209–1224. doi:10.1007/s00018-015-2117-6
12. Heuckmann JM, Balke-Want H, Malchers F, et al. Differential protein stability and ALK inhibitor sensitivity of EML4-ALK fusion variants. Clin Cancer Res. 2012;18(17):4682–4690. doi:10.1158/1078-0432.CCR-11-3260
13. Sabir SR, Yeoh S, Jackson G, Bayliss R. EML4-ALK variants: biological and molecular properties, and the implications for patients. Cancers. 2017;9(9):118. doi:10.3390/cancers9090118
14. Du X, Shao Y, Gao H, et al. CMTR1-ALK: an ALK fusion in a patient with no response to ALK inhibitor crizotinib. Cancer Biol Ther. 2018;19(11):962–966. doi:10.1080/15384047.2018.1480282
15. Yang Y, Qin SK, Zhu J, et al. A rare STRN-ALK fusion in lung adenocarcinoma identified using next-generation sequencing-based circulating tumor DNA profiling exhibits excellent response to crizotinib. Mayo Clin Proc. 2017;1(1):111–116. doi:10.1016/j.mayocpiqo.2017.04.003
16. Zhang M, Wang Q, Ding Y, et al. CUX1-ALK, a novel ALK rearrangement that responds to crizotinib in non-small cell lung cancer. J Thorac Oncol. 2018;13(11):1792–1797. doi:10.1016/j.jtho.2018.07.008
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.