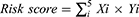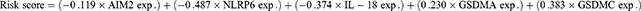Back to Journals » International Journal of General Medicine » Volume 15
Identification of a Novel Pyroptosis-Related Gene Signature Indicative of Disease Prognosis and Treatment Response in Skin Cutaneous Melanoma
Authors Li AA, Zhang Y, Tong WL, Chen JW, Huang SH, Liu JM , Liu ZL
Received 26 March 2022
Accepted for publication 22 June 2022
Published 12 July 2022 Volume 2022:15 Pages 6145—6163
DOI https://doi.org/10.2147/IJGM.S367693
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
An-An Li,1,2 Yu Zhang,1,2 Wei-Lai Tong,1,2 Jiang-Wei Chen,1 Shan-Hu Huang,1 Jia-Ming Liu,1 Zhi-Li Liu1,2
1Department of Orthopedic Surgery, The First Affiliated Hospital of Nanchang University, Nanchang, People’s Republic of China; 2Medical Innovation Center, The First Affiliated Hospital of Nanchang University, Nanchang, People’s Republic of China
Correspondence: Zhi-Li Liu; Jia-Ming Liu, Department of Orthopedic Surgery, The First Affiliated Hospital of Nanchang University, No. 17 Yongwaizheng Street, Donghu District, Nanchang, 330006, People’s Republic of China, Tel/Fax +86-791-88693201, Email [email protected]; [email protected]; [email protected]
Purpose: Pyroptosis plays an important role in the occurrence and progression of many tumors; however, the specific mechanisms involved remain unknown. Here, we construct a pyroptosis-related gene signature that can be used to predict survival prognosis of skin cutaneous melanoma (SKCM) and provide guidance for clinical treatment.
Methods: By integrating data from the two databases from the GTEx and TCGA, differentially expressed genes (DEGs) from normal tissues and skin cutaneous tumor tissues were identified. The main signaling pathways and function enrichment of these differential genes were determined. Univariate and multivariate COX regression analysis, and risk score analysis were used to construct a signature to assess its predictive value for overall survival. The mRNA expression of these five genes in melanoma cells was determined by quantitative polymerase chain reaction (qPCR). The pRRophetic algorithm was used to estimate the half-maximal inhibitory concentration (IC50) of chemotherapy drugs in SKCM patients. The expression of multiple immune checkpoint genes also was evaluated.
Results: Sixteen DEGs associated with pyroptosis in SKCM and normal skin tissues were identified. Of these, 12 pyroptosis-related DEGs were associated with the prognosis of SKCM. A five-gene signature (GSDMA, GSDMC, IL-18, NLRP6, and AIM2) model was constructed. Patients were divided into high-risk and low-risk groups using the risk scores. Of these, the high-risk group had a worse survival prognosis. There are significant differences in the predicted sensitivity of the high-risk and low-risk groups to chemotherapeutic drugs. In addition, compared with the high-risk group, the low-risk group showed higher expression of PD-1, PDL-1, CTLA-4, LAG-3, and VSIR.
Conclusion: In this study, we constructed a novel prognostic pyroptosis-related gene-signature for SKCM. These genes showed good predictive value for patient prognosis and could provide guidance for better treatment of SKCM patients.
Keywords: skin cutaneous melanoma, pyroptosis, prognosis, signature, risk model
Introductions
Skin cutaneous melanoma (SKCM) is very aggressive and lethal type of skin cancer.1 Although melanoma only accounts for about 1% of all skin cancers, the mortality rate is very high, accounting for 90% of all skin cancer deaths.2 In recent years, the incidence of melanoma has gradually increased, and ultraviolet radiation is one of the main environmental risk factors. Melanoma has a tendency to metastasize in the early stages of disease progression, and even small primary tumors may occur.3 The most common metastatic sites are lung, liver, brain, and bone.4 The treatment of SKCM is still based on surgery, adjuvant immunotherapy, and chemotherapy. Although patients with early melanoma have a good prognosis, patients with distant metastases experience a very poor prognosis, with a median survival time of only 6–9 months.5
Pyroptosis is an inflammatory form of programmed cell death. It was first described in 1992 by A. Zychlinsky in Shigella flexneri-infected macrophages. Unlike apoptosis, pyroptosis is characterized by the release of pro-inflammatory factors and relies on caspase-1 instead of caspase-3, as in apoptosis. Pyroptosis can be induced by the typical caspase-1 inflammasome pathway.6,7 As the executor of pyroptosis, Gasdermin D (GSDMD) is cleaved by activated caspase-1 into two fragments: the N-terminal domain and the C-terminal domain. The N-terminal fragment shifts to the inner lobe of the plasma membrane, forming a membrane pore with an inner diameter of 10–15 nm.8,9 Next, the punched membrane further promotes the release of inflammatory factors, causing cell swelling, membrane rupture, and ultimately leading to pyroptosis. Pyroptosis was originally discovered because of its anti-inflammatory effects. Currently, an increasing number of studies have shown that Pyroptosis also plays an important role in the immune microenvironment of tumors. Inflammatory vesicles, Gasdermin family proteins, and pro-inflammatory cytokines such as interleukin (IL)-18 are the key components of pyroptosis and are related to tumor occurrence, invasion, and metastasis.10
At present, a few studies have investigated the prognostic role of genes associated with pyroptosis in tumors from public databases. Ye et al found that seven pyroptosis-related genes could be used to predict the survival of patients with ovarian cancer.11 However, no study has found a correlation between SKCM and pyroptosis. Herein, we constructed a five-pyroptosis gene-signature related to the prognosis of patients with SKCM by combining two datasets extracted from the GTEx and TCGA databases. In addition, we analyzed the gene characteristics in the tumor microenvironment by determining the degree of immune cell infiltration, and analyzed the differences in the efficacy of different chemotherapy and immunotherapy agents in the high- and low-risk groups of SKCM patients.
Methods
Data Acquisition
RNA sequencing (RNA-seq) data of 471 SKCM patients and the corresponding clinical features were obtained from TCGA database (https://portal.gdc.cancer.gov/repository). The RNA-seq data of 813 normal human skin samples of the GTEx database were downloaded from UCSC.Xena website.(https://xenabrowser.net/datapages/).
Acquisition of Differentially Expressed Pyroptosis-Related Genes, Functional Enrichment Analysis, and Construction of PPI Network
In total, 33 pyroptosis-related genes were obtained from previous studies11 (Table 1). The expression data in both GTEx and TCGA datasets were normalized to fragment per kilobase million (FPKM) values. Then, differentially expressed genes (DEGs) of pyroptosis between normal tissues and melanoma tumors were obtained through WilcoxTest. The 16 DEGs of pyroptosis-related genes were screened using the following criteria: fold change (FC) >1, P<0.05. In order to explore the potential pathways of the 16 DEGs related to pyroptosis, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis by applying the “clusterprofiler” package. At the same time, we applied “consensusclusterplus” software package carried out consensus cluster analysis on 471 patients with SKCM. Furthermore, we also used the STRING search tool (https://string-db.org/) to build the PPI network for the identified DEGS.
 |
Table 1 List of 33 Pyroptosis-Related Genes |
Screening of Prognostic Pyroptosis-Related Signature Genes and Construction of the Risk Model
We used univariate Cox regression analysis to identify 12 key prognostic pyroptosis-related genes from the DEGs. Next, the 12 genes were selected to multivariate optimization Cox regression analysis, and finally five pyroptosis-related signature genes (GSDMA, GSDMC, AIM2, IL-18, and NLRP6) were used to construct the prognostic model for further evaluation. We used the “Scale” function in the R software to centrally normalize the TCGA expression data and calculate the risk score. The risk score formula was calculated as follows:
where, X was a gene expression coefficient, and Y indicated the gene expression level. According to the cut-off point of median risk score, SKCM patients were divided into low-risk subgroups and high-risk subgroups. Kaplan–Meier analysis was used to compare the overall survival (OS) of the two groups.
Validation of mRNA Expression of DEGs by Quantitative Polymerase Chain Reaction
The level of mRNA expression of five DEGs (GSDMA, GSDMC, AIM2, IL-18 and NLRP6) in three melanoma cell lines (A375, HS294T and M14) and in the normal HaCaT skin epithelial cell line was verified by quantitative polymerase chain reaction (qPCR).
Immunohistochemistry and Immune Cell Infiltration of the DEGs
The abundance of immune infiltrates in SKCM and the gene expression of the DEGs identified above of the immune infiltrates (GSDMA, GSDMC, AIM2, NLRP6 and IL-18) was determined using the Tumor Immune Estimation Resource (TIMER) database (https://cistrome.shinyapps.io/timer/). Further, we obtained the immunohistochemical (IHC) staining data of the protein products relative to the DEGs in SKCM from the Human Protein Atlas (HPA) database (https://www.proteinatlas.org/).
Validation of the Pyroptosis-Related Signature Gene and Construction of a Nomogram
We used univariate and multivariate Cox regression analysis to evaluate the independent prognostic value of pyroptosis-related gene risk characteristics for OS. The overall score was determined including age, sex, stage, TNM, classification, and risk score. The probability of survival at 1, 2, and 3 years was determined by constructing a nomogram, and we evaluated the performance of the nomogram by drawing ROC curves.
Exploration of the Significance of the Model in Predicting Response to Chemotherapy, Targeted Therapy, and Immunotherapy
We calculated the half-inhibitory concentration (IC50) of common chemotherapy and targeted drugs, such as cisplatin, paclitaxel, docetaxel, sorafenib, and PD0325901 (a MEK inhibitor) to evaluate the model that predicts the clinical response of SKCM treatment. Using Wilcoxon signed-rank test to compare the differences in the IC50 between the high-risk and low-risk groups. The results obtained were shown as box drawings using with pRRophetic and ggplot2 packages in R software. We used the ggstatsplot package of R software to study the relationship between the model and the expression levels of immune checkpoint-related genes, including PD-1, PDL-1, CTLA-4, LAG-3, and VSIR, and the results are presented in a violin chart.
Cell Culture
Human immortalized keratinocytes (HacaT) and three human skin melanoma cell lines (A375, HS294T and M14) were purchased from Cell Resource Center, PMUC (Beijing China). A375, HS294T and M14 cell was maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, United States) supplemented with 10% fetal bovine serum (FBS; Gibco), 1% penicillin and streptomycin (Gibco). Hacat was maintained in modified Eagle’s medium (MEM; Gibco, United States) supplemented with 15% fetal bovine serum (FBS; Gibco). All cells were cultured in a 5% CO2-humidified atmosphere at 37◦C.
Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
Using the TRIzol reagent (Invitrogen) to extracted the total RNA.The qPCR assay was performed by LightCycler480 system (Roche, Switzerland) and SYBR Green (Takara). NLRP6: Primer name(F):AGGCAATGACTGACCCACTG, Primer name(R):CTGAGCCTGTTGTGGAGGAG. AIM2: Primer name(F): TTTCAGAA GCGCTGTTTGCC, Primer name(R):TCTCCTGCTTGCCTTCTTGG. IL18: Primer name(F):TGACCAAGGAAATCGGCCTC, Primer name(R):ATGGTCCGGGGTGC ATTATC. GSDMA:Primer name(F):GTGGGCGCCATCCTCTATTT, Primer name(R):TGTTCCATCGTGCTCTCCAC. GSDMC:Primer name(F):GTGGTGAC AGAGGCTGTTGA, Primer name(R):CTCTCTCCTTGGCCTTGACC.
Statistical Analysis
One-way analysis of variance was used to compare gene expression levels in normal skin epithelial tissues and SKCM tissues. Pearson’s chi-square test was used to compare categorical variables. The Kaplan–Meier method and the two-sided Log rank test were used to analyze OS. Univariate and multivariate Cox regression models were used to evaluate the independent prognostic value of risk models. R Statistical Software version 3.6.3 and strawberry-perl-5.32.0.1 were used to analyze all data. Statistical significance was defined by differences with a P-value <0.05.
Results
DEGs Identified Between Normal and Tumor Tissues and Functional Enrichment Analysis
We compared the expression levels of 33 pyroptosis-related genes (Table 1) in the mixed Gene-Tissue Expression (GTEx) and The Cancer Genome Atlas (TCGA) data of 813 normal tissues and 471 tumor tissues, and identified 16 DEGs (all P<0.001). Among these, nine genes (ELANE, GSDMC, IL-18, GSDMA, NLRP1, GSDMB, IL6, PJVK, and CASP4) were downregulated while seven genes (IL1B, NLRP3, CASP5, NLRC4, NLRP6, AIM2, and NLRP7) were upregulated in the tumor group. The RNA expression levels of these 16 DEGs are presented as a heatmap in Figure 1A, as a volcano-map in Figure 1B, and as a bar plot in Figure 1C (Green: represents low expression; Red: represents high expression). We further analyzed the relationship between DEGs and PPI. And the results show that 13 of the 16 DEGs encode proteins that are related to each other (Figure 1D). In the PPI analysis, the minimum interaction score was set to 0.7.
GO and KEGG pathway enrichment analyses were performed to evaluate the possible functions of the 16 DEGs. The results from GO enrichment analysis indicated that the DEGs mapped to inflammasome-related GO terms, including pyroptosis, IL-1 beta production, IL-1 production, IL-1 beta secretion, chemokine biosynthetic processes, chemokine metabolic processes, IL-1 secretion, positive regulation of T cell cytokine production, positive regulation of cytokine production, and positive regulation of cysteine-type endopeptidase activity (Figure 1E). The results of the KEGG enrichment analysis identified enrichment in genes associated with the NOD-like receptor signaling pathway, Salmonella infection, Yersinia infection, the cytosolic DNA-sensing pathway, pathogenic Escherichia coli infection, and Shigellosis (Figure 1F). These results suggested that these genes may function as inflammasome responses of SKCM.
Tumor Classification Based on the DEGs
In order to study the relationship between the expression of the 16 pyroptosis-related DEGs and the subtypes of skin malignant melanoma (SKCM), we performed a consensus cluster analysis on all 471 SKCM patients in TCGA cohort. We found that when k=2 (meaning that 471 SKCM patients were divided into two clusters according to 16 DEGs), the intragroup correlations were the highest and the inter-group correlations were lowest (Figure 2A). The heatmap (Figure 2B) showed the gene expression profile and clinical characteristics, including sex (male or female), tumor progression grade (T1–T4), age (≤65 years or >65 years), distant metastasis status (M0 or M1), and lymph nodes metastasis (N0–N3). There was a significant difference in the OS between the two groups (P<0.001) (Figure 2C).
Construction of Pyroptosis-Related Prognostic 5-Gene Signature
In order to explore the potential prognostic value of these DEGs in the SKCM, Cox regression analysis was applied. Univariate Cox regression analysis showed that 12 DEGs (NLRP1, NLRP3, NLRP6, NLRP7, CASP4, CASP5, IL-18, AIM2, GSDMA, GSDMB, GSDMC, and NLRC4) were significantly associated with survival. From the multivariate regression optimization analysis, five genes (NLRP6, GSDMA, GSDMC, AIM2, IL-18) were selected for constructing a prognostic risk model. The coefficients of univariate Cox regression and multivariate Cox regression are presented in Tables 2 and 3. The risk score was calculated as follows:
 |
Table 2 The Hazard Ratio of Included Genes Obtained from Univariate Cox Regression Analysis |
 |
Table 3 The Coefficients of Included Genes Obtained from Multivariate Cox Regression Analysis |
Based on the median score calculated by the risk score formula, 471 patients were equally divided into low-risk and high-risk subgroups (Figure 3A). The mRNA expression of genes in SKCM patients is presented in a heatmap in Figure 3B. Patients in the high-risk group had a higher mortality than those in the low-risk group (Figure 3C). A notable difference in OS was detected between the low-risk and high-risk groups (P<0.001) (Figure 3D).
Independent Prognostic Value of the Risk Model
Through univariate regression analysis, we found that the risk score model constructed by the five-gene signature could be used as an independent prognostic factor to predict OS of SKCM patients (HR=1.946, 95% CI: 1.657–2.287) (Figure 4A). Further, the combined multivariate Cox analysis showed that the risk score was a prognostic factor for SKCM patients both in the high- and low-risk groups (HR=1.825, 95% CI: 1.535–2.169) (Figure 4B). The ROC curve was used to verify the prediction accuracy of risk scoring (AUC=0.724, Figure 4C). Drawing a heatmap of clinical characteristics, we found that there were significant differences in age, tumor grade, and tumor T staging between the high-risk and low-risk groups (Figure 4D). In addition, we also constructed a nomogram able to calculate the risk score to predict the 1–3 year survival rate of the patient (Figure 4E).
Protein Expression of Five Key Genes in SKCM Patients
We used Human Protein Atlas (HPA) database to explore the protein expression of the five key genes in SKCM. We evaluated the expression of NLRP3, AIM2, GSDMA, IL-18, and GSDMC protein in this database. Staining for CADMA and GSDMC was low, IL-18 and NLRP6 staining was intermediate, and AIM2 staining was high in tumor tissue (Figure 5A–E).
 |
Figure 5 Protein expression of five key genes in SKCM patients. (A–E) Immunohistochemical staining images from The HPA of five key genes in the SKCM tissue. |
Association Between Five Key Genes and Immune Cell Infiltration, and the Expression of 22 Kind of Immune Cells on Both the High- and Low- Risk Groups
With the help of the TIMER database, we explored the relationship between the expression of the five key DEGs and the infiltration of immune cells in SKCM. Expression of AIM2, GSDMA, GSDMC, IL-18, and NLRP6 were related to immune cell infiltration (Figure 6A–E). The expression of AIM2, IL-18, and NLRP6 was significantly positively correlated with levels of infiltrating CD8+T cells, neutrophils and dendritic cells (Figure 6A, D and E). The expression of AIM2, NLRP6, GSDMA, and IL-18 was positively correlated with the level of infiltrating CD4+ T cells (Figure 6A, B, D and E), while the expression of GSDMC was negatively correlated with the level of infiltrating CD4+ T cells (Figure 6C). We further analyzed the differential expression of 22 types of immune cell infiltrates in both the high- and low-risk groups (Figure 7).
 |
Figure 7 The differential expression of 22 kind of immune cells in both the high- and low-risk groups. |
Response of High- and Low-Risk Patients to Chemotherapy, Targeted Therapy, and Immunotherapy
The pRRophetic algorithm was used to predict the IC50 of three common chemotherapeutic agents (cisplatin, docetaxel, and paclitaxel) (Figure 8A1–C1) and two common targeting therapeutic agents (BRAF inhibitor sorafenib and MEK inhibitor PD0325901) (Figure 8D1–E1) in high-risk and low-risk patients. The results showed that compared with the high-risk group of SKCM patients, the low-risk group had higher IC50s for docetaxel and cisplatin, and lower IC50s for paclitaxel, PD0325901 and sorafenib. Therefore, we can infer that low-risk patients were more sensitive to the chemotherapy drug paclitaxel and less sensitive to cisplatin and docetaxel than high-risk SKCM patients. And compared with patients in high-risk group, patients in low-risk group were more sensitive to two targeted therapy drugs, sorafenib (BRAF inhibitor) and PD0325901 (MEK inhibitor). We also assessed the potential susceptibility to immune checkpoint inhibitors (targeting the immune checkpoint genes PD-1, PDL-1, CTLA-4, LAG-3, or VSIR) on both high risk and low risk groups. PD-1, PDL-1, CTLA-4, LAG-3, or VSIR are all lowly expressed in samples from the high-risk group (Figure 8A2–E2). These data suggest that high-risk patients are more sensitive to inhibitors of these immune detection targets.
Expression Levels of GSDMA, GSDMC, IL18, AIM2 and NLRP6
To better explain the biological function of these 5 genes in the pathogenesis and development of SKCM, we verified the difference in their expression levels by qPCR. The results showed that compared with normal skin cell HaCaT, GSDMA, GSDMC and IL18 are low expressed in three melanoma cells (A375, HS294T and M14), while NLRP6 was highly expressed (Figure 9A–D). In addition, AIM2 was highly expressed in A375 and HS294T cells compared to HahcaT, but there was no significant difference in expression in M14 (Figure 9E). These results were basically consistent with the results of our previous bioinformatics analysis.
Discussion
In this study, we obtained data from a total of 813 normal tissues and 471 skin melanomas by integrating two datasets from the GTEx and TCGA. Next, we compared the mRNA levels of 33 pyroptosis-related genes in the two groups of tissue samples, as a result, we identified 16 DEGs. Among these, nine genes were up-regulated, and the remaining seven genes were down-regulated. Through GO and KEGG enrichment analysis, these genes are found to be related to inflammation and immunity. We further divided the patients of SKCM into two groups using consensus cluster analysis, and found that there were significant differences in the clinical characteristics. Then, using univariate analysis and multivariate Cox regression analysis, we constructed a risk model with five DEGs to evaluate their prognostic value. In addition, we verified the mRNA expression levels of these five DEGs in melanoma cells through qPCR, and found that GSDMA, GSDMC, and IL-18 showed low expression, while AIM2 and NLRP6 were highly expressed, which was consistent with our predictions. Furthermore, the IHC staining results of the five DEGs (GSDMA, GSDMC, AIM2, NLRP6, and IL-18) from the HPA database were all positive.
Similar to apoptosis and autophagy, pyroptosis is also a mechanisms of PCD, which is usually mediated by the Gasdermins family and is accompanied by inflammation and an immune response.12 In recent years, several studies have closely linked pyroptosis with tumors, albeit their association is complicated. Pyroptosis can induce PCD to inhibit the occurrence and development of tumors, although pyroptosis can also promote inflammation and provide a suitable microenvironment for the growth of tumor cells.13 Studies have shown that pyroptosis-related genes are associated with the survival and prognosis of ovarian cancer.11 Further, pyroptosis-related signal pathways may also play an important role in gastric cancer and colorectal cancer.14 However, the influence of pyroptosis-related genes in SKCM and whether they are associated with survival is still unclear. In this study, we found that five pyroptosis genes (GSDMA, GSDMC, AIM2, NLRP6, IL-18) can be used to predict the OS of SKCM patients.
As a member of the Gasdermins family, GSDMA is expressed in epithelial cells in a variety of tissues, including the esophagus, lower digestive tract, skin, breast, and stomach.15 Human GSDMA adopts a self-inhibitory two-domain structure, in which the N-terminal domain is suppressed by the C-terminal domain.16,17 The N-terminal domain of GSDMA is similar to GSDMD. When the N-terminal domain of GSDMA is overexpressed, it will form punches in the plasma membrane and induce pyroptosis-like features,9 leading cells to swell and rupture. GSDMA is associated with autoimmune diseases, such as skin inflammation and hair loss,18,19 asthma,20,21 and gastric cancer. GSDMA is usually silenced in gastric cancer, and regulates the apoptosis of gastric epithelial cells.22 Saeki et al analyzed the expression of four Gasdermin family genes in esophageal cancer and gastric cancer, and found that GSDMA, GSDMC, and GSDMD were tumor suppressor genes.23 In our study, we also found that GSDMA is under-expressed in SKCM, which suggested that GSDMA may be also act as tumor suppressor gene in this context.
GSDMC is also a member of the Gasdermins family like GSDMA, but the biological functions and disease relevance are still not certain. Studies have shown that the Gasdermin-N domain expressed by GSDMC alone can signal cell death and trigger PCD.7 Hou et al found that GSDMC is specifically cleaved by caspase-8 with TNFα treatment, generating a GSDMC N-terminal domain that forms pores on the cell membrane and induces pyroptosis.24 Miguchi et al25 provided evidence that GSDMC, as an oncogene, could promote the proliferation of colorectal cancer cells by inhibiting the activation of Transforming growth factor beta receptor type II (TGFBR2). In addition, Wei et al26 also found that overexpression of GSDMC was also a prognostic factor for predicting poor outcomes in lung adenocarcinoma. Through analysis, we found that the expression of GSDMC in SKCM low-risk group and high-risk group was very low, and only a small number of patients had high GSDMC expression in the high-risk group. In addition, it was further revealed that GSDMC was under-expressed in melanoma cells by qPCR validation. This suggests that GSDMC may function as a tumor suppressor gene in SKCM.
Nod-like receptor pyrin domain-containing protein 6 (NLRP6) is a member of the Nod-Like Receptor (NLR) family of proteins,27 which play a key roles in innate immunity and host defense28,29. NLRP6 has three domains,30 namely the N-terminal pyrrole domain (PYD), nucleotide binding and oligomerization domain (NACHT), and the C-terminal leucine-rich repeat sequence (LRR). In some cases, NLRP6 can recruit the linker apoptosis-related dot-like protein (ASC) and inflammatory caspase-1 to form an inflammasome, mediating maturation and secretion of the pro-inflammatory cytokines IL-18 and IL-1β31, and causing pyroptosis. Wang et al32 confirmed that NLRP6 can inhibit the growth of gastric cancer. However, Ahmed et al33 revealed that the level of NLRP6 in colorectal cancer tissues was elevated, and could act as a diagnostic marker. Through this study, we found that NLRP6 is a low-risk marker gene, associated with a good prognosis of SKCM. NLRP6 plays different roles in different tumors, and may be related to the immune microenvironment in which the tumor is located and the different immune mechanism of NLRP6.
Absent in melanoma 2 (AIM2) is a founding member of the AIM2-like receptor (ALR) family and has been identified as a cytoplasmic sensor for dsDNA.34 By binding to exogenous dsDNA, AIM2 triggers the assembly of AIM2 inflammasomes, leading to caspase-1-mediated inflammation and cell death35–37. In addition to its role in the activation of inflammasomes, AIM2 also plays a role in many tumors. In oral squamous cell carcinoma (OSCC), AIM2 overexpression promotes OSCC cell proliferation and prevents cell apoptosis.38 In non-small cell lung cancer, AIM2 overexpression promotes the growth of cancer cells and predicts a lower survival rate of patients.39 However, the overexpression of AIM2 inhibited tumor growth in breast cancer.40 Interestingly, our study found that AIM2 has a high-expression in SKCM and is associated with a good prognosis.
IL-18 is a member of the IL-1 cytokine family and mediates inflammation downstream of NLRP3 and NLRP1 inflammation. At the same time, it can stimulate innate lymphocytes and T cells with antigen experience (but not naïve T cells) to trigger an immune response.41 IL-18 is not only a key regulator of signaling pathways in a variety of immune responses but also participates in the anti-tumor process of a variety of tumors. Recent studies have found that IL-18 shows an abnormally low expression in esophageal cancer and OSCC.42,43 We have confirmed that IL-18 also has a low expression in SKCM through bioinformatics prediction analysis and qPCR verification, and was enriched in the low-risk group, which indicated that IL-18 may play a role in SKCM as a tumor suppressor gene.
In summary, three genes (NLRP6, IL-18, AIM2) in our prediction model are promoters of pyroptosis, and two genes (GSDMC and GSDMA) are executor genes of pyroptosis. Pyroptosis, an inflammatory death, is considered another form of PCD. However, to date, studies on pyroptosis are limited. By constructing a risk model, we compared the high-risk and low-risk groups in SKCM and found that five DEGs mainly involved in immune response and inflammatory response and on GO enrichment analysis, these DEGs were related to pyroptosis. Further, KEGG analysis showed that the DEGs were involved in immune and inflammatory pathways. It is reasonable to speculate that pyroptosis may induce immune and inflammatory responses in the tumor microenvironment to inhibit tumor generation and metastasis. With the help of the TIMER database, we found that the expression of five DEGs were related to a variety of immune cells in SKCM. Our further analysis showed that the levels of naïve B cells, memory B cells, and activated NK cells were higher in the low-risk group, which is a beneficial anti-tumor effect. As the management of SKCM currently focuses on immune and chemotherapy treatment, we also analyzed differences in chemotherapy, targeted therapy, and immunotherapy between the high and low-risk groups of melanoma patients. The results showed that the sensitivity of the two groups of patients to cisplatin, paclitaxel, and sorafenib was significantly different. At the same time, the expression of the immune checkpoint molecules PD-1, PDL-1, CTLA-4, LAG-3, and VSIR were also significantly different between the high-risk and low-risk groups. It shows that low-risk early SKCM patients are very sensitive to chemotherapy and immunotherapy.
Here we found that pyroptosis is related to SKCM, and the expression of most of the pyrolysis-related genes was significantly different comparing normal and melanoma cells. This study provides five novel genetic markers for predicting the prognosis of SKCM patients, and provides an important basis for further research on the relationship between pyroptosis-related genes and immune function in SKCM patients. At the same time, our findings can help clinicians provide an optimal treatment plan for SKCM patients. However, our data source is relatively limited, and we did not explored the molecular mechanisms of the signature genes. Therefore, it is essential to study the specific mechanisms of the identified pyroptosis-related genes in SKCM.
Conclusions
We identified a novel prognostic pyroptosis-related gene expression signature (GSDMA, GSDMC, NLRP6, AIM2, and IL-18) in SKCM. Patient stratification based on this genetic signature can be used to improve prognostic outcomes and guide better chemotherapy and immunotherapy for SKCM patients.
Abbreviations
SKCM, skin cutaneous melanoma; DEGs, differentially expressed genes; qPCR, quantitative polymerase chain reaction; IC50, half-maximal inhibitory concentration; FPKM, fragment per kilobase million; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; TIMER, Tumor Immune Estimation Resource; HPA, Human Protein Atlas; GTEx, Gene-Tissue Expression; TCGA, The Cancer Genome Atlas.
Data Sharing Statement
The datasets analyzed in this study can be found in public online databases. Raw counts for RNA-seq transcriptome data and corresponding clinical data for skin cutaneous melanoma were extracted from TCGA (https://portal.gdc.cancer.gov) and GTEx (https://xenabrowser.net/datapages/). Pyroptosis-related genes were extracted from previous literature reports (Ye Y, Dai Q, Qi H. A novel defined pyroptosis-related gene signature for predicting the prognosis of ovarian cancer. Cell Death Discov. 2021;7:71). Infiltration data for immune cells were downloaded from the TIMER (https://cistrome.shinyapps.io/timer/) databases. Immunohistochemical staining data from Human Protein Atlas (HPA) database (https://www.proteinatlas.org/). And qPCR results were included in the article.
Ethics Approval and Consent to Participate
The data in our study are mainly from public data and do not involve ethical issues. Our study has been approved by the Medical Ethics Committee of the First Affiliated Hospital of Nanchang University.
Consent for Publication
Our manuscript contains not any individual data in any form.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas. An-An Li and Yu Zhang contributed equally to this work and share the first authorship. Zhi-li Liu and Jia-Ming Liu share the corresponding author responsibilities. All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Postgraduate Innovation Special Fund Project of Jiangxi Province, China (No.YC2021-B051).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Leiter U, Garbe C. Epidemiology of melanoma and nonmelanoma skin cancer—the role of sunlight. In: Reichrath J, editor. Sunlight, Vitamin D and Skin Cancer. Advances in Experimental Medicine and Biology. Vol. 624. New York: Springer; 2008: 89–103. doi:10.1007/978-0-387-77574-6_8
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA a Cancer J Clin. 2020;70:7–30. doi:10.3322/caac.21590
3. Bedrosian I, Faries MB, Iv DG, et al. Incidence of sentinel node metastasis in patients with thin primary melanoma (#1 mm) with vertical growth phase. Ann Surg Oncol. 2000;7:6. doi:10.1007/s10434-000-0262-z
4. Damsky WE, Rosenbaum LE, Bosenberg M. Decoding melanoma metastasis. Cancers. 2010;3:126–163. doi:10.3390/cancers3010126
5. Lens M. Current clinical overview of cutaneous melanoma. Br J Nurs. 2008;17:300–305. doi:10.12968/bjon.2008.17.5.28825
6. Liu X, Lieberman J. A mechanistic understanding of pyroptosis: the fiery death triggered by invasive infection. Adv Immunol. 2017;135:81–117.
7. Shi J, Zhao Y, Wang K, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi:10.1038/nature15514
8. Liu X, Zhang Z, Ruan J, et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535:153–158. doi:10.1038/nature18629
9. Ding J, Wang K, Liu W, et al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 2016;535:111–116. doi:10.1038/nature18590
10. Kolb R, Liu G-H, Janowski AM, Sutterwala FS, Zhang W. Inflammasomes in cancer: a double-edged sword. Protein Cell. 2014;5:12–20. doi:10.1007/s13238-013-0001-4
11. Ye Y, Dai Q, Qi H. A novel defined pyroptosis-related gene signature for predicting the prognosis of ovarian cancer. Cell Death Discov. 2021;7:71. doi:10.1038/s41420-021-00451-x
12. Shi J, Gao W, Shao F. Pyroptosis: gasdermin-mediated programmed necrotic cell death. Trends Biochem Sci. 2017;42:245–254. doi:10.1016/j.tibs.2016.10.004
13. Xia X, Wang X, Cheng Z, et al. The role of pyroptosis in cancer: pro-cancer or pro-“host”? Cell Death Dis. 2019;10:650. doi:10.1038/s41419-019-1883-8
14. Zhou C-B, Fang J-Y. The role of pyroptosis in gastrointestinal cancer and immune responses to intestinal microbial infection. Biochim Biophys Acta Rev Cancer. 2019;1872:1–10. doi:10.1016/j.bbcan.2019.05.001
15. Saeki N, Kuwahara Y, Sasaki H, Satoh H, Shiroishi T. Gasdermin (Gsdm) localizing to mouse Chromosome 11 is predominantly expressed in upper gastrointestinal tract but significantly suppressed in human gastric cancer cells. Mamm Genome. 2000;11:718–724. doi:10.1007/s003350010138
16. Lin P-H, Lin H-Y, Kuo C-C, Yang L-T. N-terminal functional domain of Gasdermin A3 regulates mitochondrial homeostasis via mitochondrial targeting. J Biomed Sci. 2015;22:44. doi:10.1186/s12929-015-0152-0
17. Ruan J, Xia S, Liu X, Lieberman J, Wu H. Cryo-EM structure of the gasdermin A3 membrane pore. Nature. 2018;557:62–67. doi:10.1038/s41586-018-0058-6
18. Kumar S, Rathkolb B, Budde BS, et al. Gsdma3I359N is a novel ENU-induced mutant mouse line for studying the function of Gasdermin A3 in the hair follicle and epidermis. J Dermatol Sci. 2012;67:190–192. doi:10.1016/j.jdermsci.2012.05.001
19. Zhou Y, Jiang X, Gu P, Chen W, Zeng X, Gao X. Gsdma3 mutation causes bulge stem cell depletion and alopecia mediated by skin inflammation. Am J Pathol. 2012;180:763–774. doi:10.1016/j.ajpath.2011.10.034
20. Ferreira MAR, Matheson MC, Tang CS, et al. Genome-wide association analysis identifies 11 risk variants associated with the asthma with hay fever phenotype. J Allergy Clin Immunol. 2014;133:1564–1571. doi:10.1016/j.jaci.2013.10.030
21. Yu J, Kang M-J, Kim B-J, et al. Polymorphisms in GSDMA and GSDMB are associated with asthma susceptibility, atopy and BHR: variants of GSDMA and GSDMB With Asthma. Pediatr Pulmonol. 2011;46:701–708. doi:10.1002/ppul.21424
22. Saeki N, Kim D, Usui T, et al. GASDERMIN, suppressed frequently in gastric cancer, is a target of LMO1 in TGF-b-dependent apoptotic signalling. Oncogene. 2007;26(45):6488–6498. doi:10.1038/sj.onc.1210475
23. Saeki N, Usui T, Aoyagi K, et al. Distinctive expression and function of four GSDM family genes (GSDMA-D) in normal and malignant upper gastrointestinal epithelium. Genes Chromosom Cancer. 2009;48:261–271. doi:10.1002/gcc.20636
24. Hou J, Zhao R, Xia W, et al. PD-L1-mediated gasdermin C expression switches apoptosis to pyroptosis in cancer cells and facilitates tumour necrosis. Nat Cell Biol. 2020;22:1264–1275. doi:10.1038/s41556-020-0575-z
25. Miguchi M, Hinoi T, Shimomura M, et al. Gasdermin C is upregulated by inactivation of transforming growth factor β receptor type II in the presence of mutated APC promoting colorectal cancer proliferation. PLoS One. 2016;11:e0166422. doi:10.1371/journal.pone.0166422
26. Wei J, Xu Z, Chen X, et al. Overexpression of GSDMC is a prognostic factor for predicting a poor outcome in lung adenocarcinoma. Mol Med Rep. 2020. doi:10.3892/mmr.2019.10837
27. Chen GY, Liu M, Wang F, Bertin J, Núñez G. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J Immunol. 2011;186:7187–7194. doi:10.4049/jimmunol.1100412
28. Anand PK, Malireddi RKS, Lukens JR, et al. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature. 2012;488:389–393. doi:10.1038/nature11250
29. Wang P, Zhu S, Yang L, et al. Nlrp6 regulates intestinal antiviral innate immunity. Science. 2015;350:826–830. doi:10.1126/science.aab3145
30. Grenier JM, Wang L, Manji GA, et al. Functional screening of five PYPAF family members identifies PYPAF5 as a novel regulator of NF-UB and caspase-1. FEBS Lett. 2002;530:6.
31. Zheng D, Kern L, Elinav E. The NLRP6 inflammasome. Immunology. 2021;162:281–289. doi:10.1111/imm.13293
32. Wang X, Wu X, Wang Q, Zhang Y, Wang C, Chen J. NLRP6 suppresses gastric cancer growth via GRP78 ubiquitination. Exp Cell Res. 2020;395:112177. doi:10.1016/j.yexcr.2020.112177
33. Ahmed FE, Vos P. Molecular markers for human colon cancer in stool and blood identified by RT-PCR. Anticancer Res. 2004;24:8.
34. Bürckstümmer T, Baumann C, Blüml S, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi:10.1038/ni.1702
35. Shrivastava G, León-Juárez M, García-Cordero J, Meza-Sánchez DE, Cedillo-Barrón L. Inflammasomes and its importance in viral infections. Immunol Res. 2016;64:1101–1117. doi:10.1007/s12026-016-8873-z
36. Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–1022. doi:10.1016/j.cell.2014.04.007
37. Fernandes-Alnemri T, Yu J-W, Juliana C, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi:10.1038/ni.1859
38. Kondo Y, Nagai K, Nakahata S, et al. Overexpression of the DNA sensor proteins, absent in melanoma 2 and interferon-inducible 16, contributes to tumorigenesis of oral squamous cell carcinoma with p53 inactivation. Cancer Sci. 2012;103:782–790. doi:10.1111/j.1349-7006.2012.02211.x
39. Zhang M, Jin C, Yang Y, et al. AIM2 promotes non‐small‐cell lung cancer cell growth through inflammasome‐dependent pathway. J Cell Physiol. 2019;234:20161–20173. doi:10.1002/jcp.28617
40. Chen I-F, Ou-Yang F, Hung J-Y, et al. AIM2 suppresses human breast cancer cell proliferation in vitro and mammary tumor growth in a mouse model. Mol Cancer Ther. 2006;5:1–7. doi:10.1158/1535-7163.MCT-05-0310
41. Zhou T, Damsky W, Weizman O-E, et al. IL-18BP is a secreted immune checkpoint and barrier to IL-18 immunotherapy. Nature. 2020;583:609–614. doi:10.1038/s41586-020-2422-6
42. Xu X, Song C, Chen Z, et al. Downregulation of HuR inhibits the progression of esophageal cancer through interleukin-18. Cancer Res Treat. 2018;50:71–87. doi:10.4143/crt.2017.013
43. Li Y, Xu Z, Li J, Ban S, Duan C, Liu W. Interleukin‐18 expression in oral squamous cell carcinoma: its role in tumor cell migration and invasion, and growth of tumor cell xenografts. FEBS Open Bio. 2018;8:1953–1963. doi:10.1002/2211-5463.12532
44. Ursini F, Maiorino M. Lipid peroxidation and ferroptosis: the role of GSH and GPx4. Free Radic Biol Med. 2020;152:175–185. doi:10.1016/j.freeradbiomed.2020.02.027
45. Carriere J, Dorfleutner A, Stehlik C. NLRP7: from inflammasome regulation to human disease. Immunology. 2021;163(4):363–376. doi:10.1111/imm.13372
46. Rossi MN, Pascarella A, Licursi V, et al. NLRP2 regulates proinflammatory and antiapoptotic responses in proximal tubular epithelial cells. Front Cell Dev Biol. 2019;7:252. doi:10.3389/fcell.2019.00252
47. Ke H, Wang X, Zhou Z, et al. Effect of weimaining on apoptosis and Caspase-3 expression in a breast cancer mouse model. J Ethnopharmacol. 2021;264:113363. doi:10.1016/j.jep.2020.113363
48. Vigneswara V, Akpan N, Berry M, et al. Combined suppression of CASP2 and CASP6 protects retinal ganglion cells from apoptosis and promotes axon regeneration through CNTF-mediated JAK/STAT signalling. Brain. 2014;137(Pt 6):1656–1675. doi:10.1093/brain/awu037
49. Chen JX, Huang XY, Wang P, et al. Effects and mechanism of arachidonic acid against TNF-α induced apoptosis of endothelial cells. Clin Hemorheol Microcirc. 2021;77(3):259–265. doi:10.3233/CH-200946
50. Xiong S, Hong Z, Huang LS, et al. IL-1β suppression of VE-cadherin transcription underlies sepsis-induced inflammatory lung injury. J Clin Invest. 2020;130(7):3684–3698. doi:10.1172/JCI136908
51. Xiao H, Li H, Wang JJ, et al. IL-18 cleavage triggers cardiac inflammation and fibrosis upon β-adrenergic insult. Eur Heart J. 2018;39(1):60–69. doi:10.1093/eurheartj/ehx261
52. Fritsch M, Günther SD, Schwarzer R, et al. Caspase-8 is the molecular switch for apoptosis, necroptosis and pyroptosis. Nature. 2019;575(7784):683–687. doi:10.1038/s41586-019-1770-6
53. Bode JG, Albrecht U, Häussinger D, et al. Hepatic acute phase proteins–regulation by IL-6- and IL-1-type cytokines involving STAT3 and its crosstalk with NF-κB-dependent signaling. Eur J Cell Biol. 2012;91(6–7):496–505. doi:10.1016/j.ejcb.2011.09.008
54. Miao H, Wang L, Zhan H, et al. A long noncoding RNA distributed in both nucleus and cytoplasm operates in the PYCARD-regulated apoptosis by coordinating the epigenetic and translational regulation. PLoS Genet. 2019;15(5):e1008144. doi:10.1371/journal.pgen.1008144
55. Trojan J, Brieger A, Raedle J, et al. BAX and caspase-5 frameshift mutations and spontaneous apoptosis in colorectal cancer with microsatellite instability. Int J Colorectal Dis. 2004;19(6):538–544. doi:10.1007/s00384-004-0597-1
56. Kumari P, Russo AJ, Shivcharan S, Rathinam VA. AIM2 in health and disease: inflammasome and beyond. Immunol Rev. 2020;297(1):83–95. doi:10.1111/imr.12903
57. Lo Verso L, Dumont K, Lessard M, et al. The administration of diets contaminated with low to intermediate doses of deoxynivalenol and supplemented with antioxidants and binding agents slightly affects the growth, antioxidant status, and vaccine response in weanling pigs. J Anim Sci. 2021;99(9):skab238. doi:10.1093/jas/skab238
58. Freeman L, Guo H, David CN, et al. NLR members NLRC4 and NLRP3 mediate sterile inflammasome activation in microglia and astrocytes. J Exp Med. 2017;214(5):1351–1370. doi:10.1084/jem.20150237
59. Sendler M, van den Brandt C, Glaubitz J, et al. NLRP3 inflammasome regulates development of systemic inflammatory response and compensatory anti-inflammatory response syndromes in mice with acute pancreatitis. Gastroenterology. 2020;158(1):253–269.e14. doi:10.1053/j.gastro.2019.09.040
60. Zamyatina A, Heine H. Lipopolysaccharide recognition in the crossroads of TLR4 and Caspase-4/11 mediated inflammatory pathways. Front Immunol. 2020;11:585146. doi:10.3389/fimmu.2020.585146
61. Tsuchiya K, Nakajima S, Hosojima S, et al. Caspase-1 initiates apoptosis in the absence of gasdermin D. Nat Commun. 2019;10(1):2091. doi:10.1038/s41467-019-09753-2
62. Thimmappa PY, Nair AS, Najar MA, et al. Quantitative phosphoproteomics reveals diverse stimuli activate distinct signaling pathways during neutrophil activation. Cell Tissue Res. 2022. doi:10.1007/s00441-022-03636-7
63. Kambara H, Liu F, Zhang X, et al. Gasdermin D exerts anti-inflammatory effects by promoting neutrophil death. Cell Rep. 2018;22(11):2924–2936. doi:10.1016/j.celrep.2018.02.067
64. Baig MS, Liu D, Muthu K, et al. Heterotrimeric complex of p38 MAPK, PKCδ, and TIRAP is required for AP1 mediated inflammatory response. Int Immunopharmacol. 2017;48:211–218. doi:10.1016/j.intimp.2017.04.028
65. Qian L, Shi H, Ding M. Comparative analysis of gene expression profiles in children with type 1 diabetes mellitus. Mol Med Rep. 2019;19(5):3989–4000. doi:10.3892/mmr.2019.10099
66. Domínguez-Ruiz M, Rodríguez-Ballesteros M, Gandía M, et al. Novel pathogenic variants in PJVK, the gene encoding pejvakin, in subjects with autosomal recessive non-syndromic hearing impairment and auditory neuropathy spectrum disorder. Genes. 2022;13(1):149. doi:10.3390/genes13010149
67. Zhou F, Li Y, Huang Y, et al. Upregulation of CASP9 through NF-κB and its target MiR-1276 contributed to TNFα-promoted apoptosis of cancer cells induced by doxorubicin. Int J Mol Sci. 2020;21(7):2290. doi:10.3390/ijms21072290
68. Zhang C, Hsu AC, Pan H, et al. Columbianadin Suppresses Lipopolysaccharide (LPS)-induced inflammation and apoptosis through the NOD1 pathway. Molecules. 2019;24(3):549. doi:10.3390/molecules24030549
69. Kang R, Zeng L, Zhu S, et al. Lipid peroxidation drives gasdermin D-mediated pyroptosis in lethal polymicrobial sepsis. Cell Host Microbe. 2018;24(1):97–108.e4. doi:10.1016/j.chom.2018.05.009
70. Zhong FL, Mamaï O, Sborgi L, et al. Germline NLRP1 mutations cause skin inflammatory and cancer susceptibility syndromes via inflammasome activation. Cell. 2016;167(1):187–202.e17. doi:10.1016/j.cell.2016.09.001
71. Li YQ, Peng JJ, Peng J, Luo XJ. The deafness gene GSDME: its involvement in cell apoptosis, secondary necrosis, and cancers. Naunyn Schmiedebergs Arch Pharmacol. 2019;392(9):1043–1048. doi:10.1007/s00210-019-01674-7
72. Schwarzer R, Jiao H, Wachsmuth L, et al. FADD and Caspase-8 regulate gut homeostasis and inflammation by controlling MLKL- and GSDMD-mediated death of intestinal epithelial cells. Immunity. 2020;52(6):978–993.e6. doi:10.1016/j.immuni.2020.04.002
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.