Back to Journals » Journal of Hepatocellular Carcinoma » Volume 9
Identification and Validation of Novel Biomarkers for Hepatocellular Carcinoma, Liver Fibrosis/Cirrhosis and Chronic Hepatitis B via Transcriptome Sequencing Technology
Authors Zhao D , Zhang X, Tang Y, Guo P, Ai R , Hou M, Wang Y, Yuan X, Cui L, Zhang Y, Zhao S, Li W, Wang Y, Sun X, Liu L, Dong S, Li L , Zhao W, Nan Y
Received 7 January 2022
Accepted for publication 27 April 2022
Published 9 May 2022 Volume 2022:9 Pages 389—403
DOI https://doi.org/10.2147/JHC.S357380
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmed Kaseb
Dandan Zhao,1,2 Xiaoxiao Zhang,1,2 Yuhui Tang,1,2 Peilin Guo,1,2 Rong Ai,1,2 Mengmeng Hou,1,2 Yiqi Wang,1,2 Xiwei Yuan,1,2 Luyao Cui,1,2 Yuguo Zhang,1,2 Suxian Zhao,1,2 Wencong Li,1,2 Yang Wang,1,2 Xiaoye Sun,1,2 Lingdi Liu,1,2 Shiming Dong,1,2 Lu Li,1,2 Wen Zhao,1,2 Yuemin Nan1,2
1Department of Traditional and Western Medical Hepatology, Third Hospital of Hebei Medical University, Shijiazhuang, Hebei, People’s Republic of China; 2Hebei Provincial Key Laboratory of Liver Fibrosis in Chronic Liver Diseases, Shijiazhuang, Hebei, People’s Republic of China
Correspondence: Yuemin Nan, Department of Traditional and Western Medical Hepatology, Third Hospital of Hebei Medical University, No. 139 Ziqiang Road, Shijiazhuang, Hebei Province, 050051, People’s Republic of China, Tel +86 311-66781227, Fax +86 311-66781289, Email [email protected]
Purpose: The aim of this study was to identify and validate novel biomarkers for distinguishing among hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC), liver fibrosis/liver cirrhosis (LF/LC) and chronic hepatitis B (CHB).
Patients and Methods: Transcriptomic sequencing was conducted on the liver tissues of 5 patients with HCC, 5 patients with LF/LC, 5 patients with CHB, and 4 healthy controls. The expression levels of selected mRNAs and proteins were assessed by quantitative real-time polymerase chain reaction (qRT-PCR) and immunohistochemical (IHC) staining, and were verified in validation set (n=200) and testing set (n=400) via enzyme-linked immunosorbent assay (ELISA).
Results: A total of 9 hub mRNAs were identified by short time-series expression miner and weighted gene co-expression network analysis. Of note, the results of qRT-PCR and IHC staining demonstrated that SHC adaptor protein 1 (SHC1), SLAM family member 8 (SLAMF8), and interleukin-32 (IL-32) exhibited gradually increasing trends in the four groups. Subsequent ELISA tests on the validation cohort indicated that the plasma levels of SHC1, SLAMF8 and IL-32 also gradually increased. Furthermore, a diagnostic model APFSSI (age, PLT, ferritin, SHC1, SLAMF8 and IL-32) was established to distinguish among CHB, LF/LC and HCC. The performance of APFSSI model for discriminating CHB from healthy subjects (AUC=0.966) was much greater compared to SHC1 (AUC=0.900), SLAMF8 (AUC=0.744) and IL-32 (AUC=0.821). When distinguishing LF/LC from CHB, APFSSI was the most outstanding diagnostic parameter (AUC=0.924), which was superior to SHC1, SLAMF8 and IL-32 (AUC=0.812, 0.684 and 0.741, respectively). Likewise, APFSSI model with the greatest AUC value displayed an excellent performance for differentiating between HCC and LF/LC than other variables (SHC1, SLAMF8 and IL-32) via ROC analysis. Finally, the results in the test set were consistent with those in the validation set.
Conclusion: SHC1, SLAMF8 and IL-32 can differentiate among patients with HCC, LF/LC, CHB and healthy controls. More importantly, the APFSSI model greatly improves the diagnostic accuracy of HBV-associated liver diseases.
Keywords: HBV-associated liver diseases, transcriptome sequencing technology, SHC adaptor protein 1, SLAM family member 8, interleukin-32
Introduction
Hepatitis B virus (HBV) infection is a global health issues, especially in developing countries, which affects 257 million people and causes 887,000 deaths annually.1 The three main stages in the progression of HBV infection are hepatocellular carcinoma (HCC), liver fibrosis/liver cirrhosis (LF/LC) and chronic hepatitis B (CHB), or widely recognized as “hepatitis trilogy”.2 CHB is commonly known for the heterogeneous patient features and long course of disease progression,3 while LF is a wound healing process. Both are necessary stages for the progression from chronic liver disease to cirrhosis, and even HCC.4 Despite great advancements in diagnosis and treatment, the majority of HCC patients have unfavorable outcomes, with a 5-year overall survival rate of <50%.5 Thus, it is of great importance to identify at-risk patients for early management of this disease.6
Currently, there are few specific clinical non-invasive diagnostic methods for HBV-related liver diseases. The gold standard for diagnosing LF or liver cancer still underlies a tissue pathology-based approach, which is known as an invasive procedure with potential risks or complications.6,7 Moreover, a number of scoring systems and morphological tests, such as FIB-4 (Fibrosis index based on the 4 factor), APRI (aspartate aminotransferase-to-platelet ratio index), and transient elastography, have been recently used to diagnose and assess liver fibrosis and cirrhosis.8 However, FIB-4 and APRI are affected by transaminases, while liver elastography is limited by ascites, morbid obesity, and small intercostal space.9 Besides, alpha-fetoprotein (AFP) is among the most common biomarkers for early detection and monitoring of HCC patients.10 However, the low accuracy and reliability of AFP have hindered its implementation into the clinic.11 Several studies have established and validated multi-parametric risk assessment models to predict the occurrence of HCC. At present, REACH-B and PAGE-B12 could predict the occurrence of HCC in CHB patients, THRI13 could predict the risk of HCC development in cirrhotic patients, aMAP score14 and GALAD model could estimate the likelihood of HCC in individual patients with chronic liver disease. However, these models have their own limitations. For early diagnosis of HCC, liquid biopsy is significantly better than conventional serological markers, but high cost can make it difficult to achieve broad applications in the clinic.15 Thus, a novel non-invasive method for diagnosing and distinguishing between HCC, LF/LC and CHB is urgently needed in order to further improve the survival outcomes of these patients.
SHC adaptor protein 1 (SHC1), a family member of adapter proteins, is consisted of p46Shc, p52Shc and p66Shc isoforms that integrate and transduce external stimuli to different signaling networks.16 SHC1 regulates reactive oxygen species (ROS) levels, cell proliferation and apoptosis, and oxidative stress.17 All the three isoforms have been proven to be associated with acute and chronic liver injuries, and play essential roles in LF18 and HCC.19 Moreover, SHC1 is linked to several other diseases, such as metabolic disorders (eg, non-alcoholic fatty liver, obesity and cardiovascular diseases),20 and cancers (eg, breast, lung, prostate and gastrointestinal cancers).21 SLAM family member 8 (SLAMF8)/CD353 is a member of the CD2 family, which reduces the production of ROS and regulates the development and function of many immune cells.22 SLAMF8 has been reported to be implicated in autoimmune inflammation,23 gastrointestinal cancers,24 and acute liver injuries.25 However, there are no previous studies examining the relationship between chronic liver diseases and SLAMF8. Interleukin-32 (IL-32) is a newly pro-inflammatory cytokine, which can induce the expression levels of TNF-α, IL-8, IL-1b, IL-6 and IFNγ. IL-32 plays a pivotal role in multiple diseases, such as infectious diseases, autoimmune diseases,26 hematologic malignancies and solid tumors.27 Furthermore, IL-32 is also upregulated in HBV-infected individuals.28,29 To sum up, we are curious about the roles of these indicators in HBV-related chronic hepatitis, liver fibrosis/cirrhosis and hepatoma.
As we all know, the mRNA expression profiles obtained from different models are varied significantly due to the different stages of HBV-related liver diseases.30 As mRNA can be translated into protein and blood sample is easily available for clinical laboratory experiments, our study aimed to verify potential mRNA biomarkers for early diagnosis and predicting the progression of HBV-associated liver diseases.31 By comparing differentially expressed mRNAs among HCC patients, LF/LC patients, CHB patients and healthy controls via transcriptome sequencing technology, some candidate targets were identified as new biomarkers for predicting the progression of HBV-associated hepatic diseases.
Materials and Methods
Study Subjects
In this cross-sectional study, we collected liver or blood samples from 180 HBV-related HCC patients, 180 HBV-associated LF/LC patients, 180 CHB patients and 180 healthy controls (CON) at the Third Hospital of Hebei Medical University between January 2017 and February 2021 according to our inclusion criteria. In accordance with the European Association for the Study of the Liver (EASL) guidelines,32 CHB was defined as chronic necroinflammatory disease of the liver caused by persistent infection with HBV. According to the American Association for the Study of Liver Diseases (AASLD) guidelines,33 LF/LC was diagnosed based on clinical examination, biochemical analysis, abdominal imaging and/or histological assessment. In compliance with the European Society of Medical Oncology (ESMO) clinical practice guidelines,34 the diagnosis of HCC was established based on histopathological confirmation, or detection of a positive lesion with recommended imaging techniques and contrast agents. At the same time, 180 age/gender-matched healthy volunteers according to the basic information of liver tissue sequencing samples, who visited our hospital for physical examination were selected as controls. Table S1 shows the inclusion and exclusion criteria of participants in this study. These subjects were randomly categorized into training, validation and testing cohorts for the identification and validation of novel biomarkers. Routine blood tests and liver biochemical tests were conducted in the local laboratories. Baseline and clinical data of the studied subjects were collected. Ethical approval for this study was obtained from the Ethics Committee of the Third Hospital of Hebei Medical University. All participants provided written informed consent prior to study enrollment. The experimental protocol was performed in compliance with the 1964 Helsinki Declaration or comparable standards.
Sample Preparation
Fresh frozen liver tissues were used for RNA-seq and PCR assay, while formalin-fixed and paraffin-embedded (FFPE) samples were used for IHC analysis. Peripheral blood mononuclear cells (PBMCs) were isolated from the whole blood of HBV-related liver disease patients and control subjects through density-gradient centrifugation method using Ficoll medium (Solarbio Life Sciences, Beijing, China). Blood samples were withdrawn from all subjects, and plasma samples were stored at ‑80°C. The plasma levels of candidate biomarkers were assessed by commercial enzyme-linked immunosorbent assay (ELISA) kits.
Transcriptomic and Bioinformatic Analyses
Transcriptome sequencing technology was conducted on HCC patients (n=5), LF/LC patients (n=5), CHB patients (n=5) and healthy controls (n=4). The detailed protocols for both transcriptomic and bioinformatic analyses are summarized in the Supplemental Methods. Briefly, Trizol DP431 reagent (Tiangen, Beijing, China) was used to isolate total RNA. Then, paired-end libraries were sequenced by Illumina Novaseq™ 6000 (LC Bio, China). A total of 332,720,614, 405,844,258, 425,724,230 and 430,992,628 reads were sequenced from CON, CHB, LF/LC, and HCC samples, respectively. For CON samples, 314,022,012 clean reads were obtained, which accounted for 94.4% of reads. Meanwhile, 384,396,180, 404,504,412 and 407,007,164 clean reads were obtained for CHB, LF/LC and HCC samples, respectively. Weighted gene co-expression network analysis (WGCNA) package in R software (https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/Tutorials/) was used to uncover the relationships among corresponding genes. Differentially expressed genes were clustered by short time-series expression miner (STEM) method (http://www.cs.cmu.edu/~jernst/stem/), and the statistically significant (P<0.05) profile boxes were coloured.
QRT-PCR Analysis
The mRNA expression of each target gene was detected by TB Green™ Premix Ex Taq™ II (Tli RNaseH Plus; Takara, Japan) using an ABI 7500 RT-PCR system (Applied Biosystems, Carlsbad, CA). Primer sequences are listed in Table 1. GAPDH expression was included as an internal control.35 The detailed method is described in the Supporting Information.
 |
Table 1 The Primer Sequences (Human) |
IHC Staining
IHC was carried out according to a previous method.35 First, the sections were dewaxed, dehydrated and blocked with blocking serum. Next, the sections were exposed to the antibody against SHC (Catalog # CY7157, 1:100, Abways), SLAMF8 (Catalog # TA349714, 1:100, Origene), IL-32 (Catalog # 11079-1-AP, 1:100, Proteintech), integrin subunit beta 2 (Catalog # 10554-1-AP, ITGB2/CD18, 1:200, Proteintech) or mesencephalic astrocyte-derived neurotrophic factor (Catalog # 10869-1-AP, MANF/ARMET, 1:200, Proteintech). After that, the sections were exposed to the HRP-conjugated secondary antibody (Proteintech) for 1 h. Lastly, all sections were visualized with 3,3’-diaminobenzidine, and counterstained with hematoxylin. Integral optical density (IOD) was recorded at ×400 magnification in 10 non-overlapping fields per sample using an Image-Pro Plus v6.0 software.
Statistical Analysis
SPSS v26.0, MedCalc v15.0 and GraphPad Prism v8.0 software were employed to perform the statistical tests. Continuous variables were presented as medians (interquartile ranges) or means ± standard deviations. Kruskal–Wallis H-test or one-way ANOVA was used to compare the differences between groups. Frequency data were shown as numbers and percentages, and were analyzed with the Chi-squared test. Independent diagnostic factors for HBV-associated liver diseases were identified by multivariate ordinal logistic regression analysis. The best threshold/cut-off value for the relative expression of each candidate biomarker in HCC, LF/LC and CHB patients were determined by ROC curve analysis using the MedCalc software. All tests were two-tailed, and a P-value of <0.05 was deemed statistically significant.
Results
Identification of Candidate Biomarkers for the Progression of HBV-Related Diseases
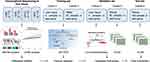
Figure 1 shows the overall design of the study. Bioinformatic analysis was then performed on all sequenced mRNAs to identify the key biomarkers in the four groups. Compared with the CON group, 1581 differentially expressed mRNAs, including 901 upregulated and 680 downregulated, were detected in the CHB group; 2141 differential expressed mRNAs, including 1493 upregulated and 648 downregulated in the LF/LC group; and 1588 differentially expressed mRNAs, including 1130 upregulated and 458 downregulated mRNAs, in the HCC group (Figure S1A). Then, we used Venn and Volcano maps to analyze differentially expressed mRNAs (Figure S1B–E). To determine the key genes and modules involved in HBV-related liver diseases, WGCNA was conducted to reveal the highly correlated gene pairs as well as the co-expression networks. As shown in Figure 2A–C, the power of β was fixed at 6 to establish a scale-free network, and the hierarchical clustering dendrogram identified 30 gene modules. As we were interested in the potential biomarkers that could be practical and convenient for predicting the progression of HBV-associated liver diseases, the green-yellow, green, turquoise and white modules were screened out. Subsequently, differentially expressed mRNAs were clustered by STEM method, and the results demonstrated that 25 mRNAs were highly expressed among CON, CHB, LF/LC and HCC groups (Figure 2D and E). By combining with WGCNA analysis, 9 progressively overexpressed mRNAs, such as SHC1, SLAMF8, IL-32, ITGB2, MANF, tandem C2 domains, nuclear (TC2N), synaptopodin (SYNPO), UDP-glucose pyrophosphorylase 2 (UGP2), EGF-containing fibulin extracellular matrix protein 1 (EFEMP1), were chosen for further analyses. KEGG pathway and GO enrichment analyses on these 9 mRNAs indicated that the functions were primarily related to natural killer cell mediated cytotoxicity, protein binding, and metal ion binding (Figure 2F and G).
Verification of Progressively Elevated mRNA Abundance Through qRT-PCR
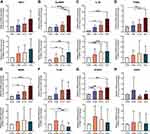
The expression levels of 9 progressively elevated mRNAs were verified with qRT-PCR in liver tissues (n=5 per group). Of these, 8 mRNAs (SHC1, SLAMF8, IL-32, ITGB2, MANF, TC2N, SYNPO and EFEMP1) showed a gradually increasing trend in the four groups (Figure 3). We further performed qRT-PCR on PBMCs (n=20 per group), and 3 mRNAs (SHC1, SLAMF8 and IL-32) had the same expression trends as those observed in liver tissues (Figure 3). Our qRT-PCR data showed that EFEMP1 has an increasing trend in liver tissues, but its expression in PBMCs is too low to be detected. Recent studies have shown that ITGB2 is elevated in many malignant tumors,36,37 and Zhang et al38 demonstrated that the expression of ITGA2 was higher in liver cancer tissues than those in para-carcinoma tissues, and it had a close relationship with ITGB2. However, it remains unknown whether the expression of ITGB2 is elevated in hepatocellular carcinoma. He et al39 showed that MANF was overexpressed in HCC, but Liu et al40 found that the mRNA and protein levels of MANF were lower in HCC tissues than in adjacent non-cancer tissues. Thus, we hypothesize that the differential expression of MANF is associated with liver cancer. To sum up, five candidate mRNAs (SHC1, SLAMF8, IL-32, ITGB2 and MANF) were selected for further verification.
IHC Staining for SHC1, SLAMF8, IL-32, ITGB2 and MANF in Normal and Pathologically Altered Livers
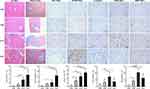
The expression activities of SHC1, SLAMF8, IL-32, ITGB2 and MANF in liver tissues were detected by IHC staining (n=5 per group). With an increase in the severity of HBV-related liver diseases, the IHC intensities of SHC1 and SLAMF8 were gradually elevated (Figure 4). IOD analysis indicated that the hepatic expression of IL-32 was the highest in HCC patients, followed by LF/LC patients, CHB patients and control subjects (Figure 4). Besides, the expression activities of ITGB2 and MANF were markedly higher in LF/LC tissues than in CHB and healthy tissues. However, ITGB2 and MANF levels were found to be declined in the HCC tissues when compared to those in LF/LC tissues (Figure 4).
Preliminary Validation of the Candidate Biomarkers via ELISA
As shown in Table 2, the proportion of male participants, age, aspartate transaminase (AST), ferritin, FIB-4 and APRI levels were markedly increased among the four groups, whereas the platelet count (PLT), white blood cell count (WBC) and albumin (ALB) levels were reduced. Notably, the plasma levels of SHC1 were remarkably elevated in HCC patients compared to LF/LC patients, CHB patients and control subjects (10.7±2.0 vs 9.7±1.7, 7.5±1.7 and 4.6±1.3 ng/mL, respectively, P<0.001; Table 2). As the disease progressed, the median values of SLAMF8 were obviously increased compared with those measured previously (P<0.001; Table 2), which were 8.8, 6.9, 6.1 and 4.9 ng/mL in the HCC, LF/LC, CHB and healthy control groups, respectively. The mean concentrations of IL-32 were 78.3±17.5 pg/mL for HCC samples, 62.3±16.0 pg/mL for LF/LC samples, 49.1±13.8 pg/mL for CHB samples and 32.3±11.7 pg/mL for control samples, which were significantly different among these groups (P<0.001; Table 2).
 |
Table 2 Clinical and Laboratory Characteristics of the Participants in the Validation Set |
Verifying the Predictive mRNA Panel in the Validation Set
Ten variables (gender, age, WBC, PLT, ALB, AST, ferritin, SHC1, SLAMF8, and IL-32) were identified by univariate ordinal regression analysis (all P<0.001; Table 3). These variables were further subjected to multivariate ordinal regression analysis, and significant differences were observed for the following variables: age (OR=1.071; 95% CI=1.036–1.107; P<0.001), PLT (OR=0.988; 95% CI=0.981–0.995; P=0.002), ferritin (OR=1.003; 95% CI=1.001–1.006; P=0.014), SHC1 (OR=2.077; 95% CI=1.648–2.618; P<0.001), SLAMF8 (OR=2.104; 95% CI=1.583–2.796; P<0.001), and IL-32 (OR=1.054; 95% CI=1.025–1.083; P<0.001).
 |
Table 3 Uni- and Multi-Variate Ordinal Logistic Analyses for the Variables Extracted by Forward Selection in the Validation Set |
When differentiating between CHB and healthy control, the AUC values of age, PLT, ferritin, SHC1, SLAMF8 and IL-32 were 0.744, 0.699, 0.688, 0.900, 0.744 and 0.821, respectively (Table 4; Figure 5). Accordingly, the cut-off values of age, PLT, ferritin, SHC1, SLAMF8 and IL-32 were 36 years old, 195×109/L, 90.60 ng/mL, 5.03 ng/mL, 4.94 ng/mL and 48.29 pg/mL, respectively. When discriminating CHB patients from LF/LC patients, age alone yielded an AUC of 0.592 with 58.0% sensitivity and 62.0% specificity at the threshold of 42 years old; PLT alone yielded an AUC of 0.812 with 66.0% sensitivity and 88.0% specificity at the threshold of 140×109/L; ferritin alone yielded an AUC of 0.587 with 54.0% sensitivity and 72.0% specificity at the threshold of 126.30 ng/mL; SHC1 alone yielded an AUC of 0.812 with 82.0% sensitivity and 64.0% specificity at the threshold of 8.11 ng/mL; SLAMF8 alone yielded an AUC of 0.684 with 44.0% sensitivity and 92.0% specificity at the threshold of 7.30 ng/mL; IL-32 alone yielded an AUC of 0.741 with 48.0% sensitivity and 90.0% specificity at the threshold of 62.37 pg/mL (Table 4; Figure 5). When distinguishing HCC patients from LF/LC patients, SLAMF8 was the most outstanding diagnostic parameter in the validation set (AUC=0.802), which was superior to age, PLT, ferritin, SHC1 and IL-32 (AUC=0.758, 0.625, 0.636, 0.646 and 0.761, respectively).
 |
Table 4 Comparison of Diagnostic Performance for Age, PLT, Ferritin, Plasma SHC1, SLAMF8, IL-32 and New Model for Patients with Different Stages of Disease |
Furthermore, a logistic regression model was constructed to determine the best diagnostic mRNA panel. The predicted probability of the 3-mRNAs panel in combination with clinical parameters was calculated as follows: APFSSI = −0.1871 + 0.012 × age - 0.0027 × PLT + 0.0005 × ferritin + 0.1361 × SHC1 + 0.1234 × SLAMF8 + 0.0108 × IL-32. At the threshold of 1.624, the sensitivity and specificity of APFSSI for CHB diagnosis were 92.0% and 94.0% with an AUC of 0.966. Using an APFSSI value of 2.470 as the decision threshold, LF/LC patients could be differentiated from CHB patients with 90.0% sensitivity and 84.0% specificity. At a score cut-off of 3.349, the diagnostic model exhibited 84.0% sensitivity and 86.0% specificity for distinguishing HCC patients from LF/LC patients (Table 4; Figure 5).
Establishing the mRNA Panel in the Test Set
Parameters estimated from the validation set (n=200) were used to evaluate the diagnostic performance of the mRNA panel in the independent test set (n=400). The variables with gradually increasing or decreasing trends were consistent with those in the validation set (Supplemental Table 2). Moreover, the mRNA panel with the highest AUCs showed a better diagnosis for HCC, LF/LC or CHB than other parameters (all P<0.05; Supplemental Table 3, Figure S2). In differentiating CHB from healthy controls, SLAMF8 was regarded as an excellent diagnostic parameter (AUC=0.911), but still inferior to APFSSI model (AUC=0.980) (Supplemental Table 3; Figure S2). The mRNA panel had a great diagnostic potential for distinguishing LF/LC patients from CHB patients, with 94.0% sensitivity and 90.0% specificity. The differentiation power of the APFSSI model for HCC vs LF/LC (AUC=0.930) was far superior to SHC1 (AUC=0.744), SLAMF8 (AUC=0.791) and IL-32 (AUC=0.852) (Supplemental Table 3; Figure S2).
Discussion
In the present study, we developed a clinically convenient and feasible non-invasive biomarker panel to help detect the potential risks of HCC, LF/LC and CHB via transcriptome sequencing.41 The primary outcomes of this study are as follows. Firstly, transcriptomic analysis identified 9 progressively elevated mRNAs during the progression of HBV-related liver injuries. Secondly, the integrated analyses of qRT-PCR and IHC demonstrated that SHC1, SLAMF8 and IL-32 were highly expressed in both liver tissues and PBMCs throughout the courses of HBV-associated liver diseases. Thirdly, we observed significant increases in the plasma levels of SHC1, SLAMF8 and IL-32 in HBV-associated liver diseases compared to healthy controls, and these mRNAs showed gradually increasing trends from CHB to LF/LC and HCC. Fourthly, when analyzed in conjunction with the clinical parameters, SHC1, SLAMF8 and IL-32 remained as independent diagnostic factors for CHB, LF/LC and HCC in a multivariate ordinal regression analysis. Lastly, it was found that the APFSSI model exhibited high diagnostic accuracy in differentiating CHB from healthy subjects, LF/LC patients from CHB patients, and HCC patients from LF/LC patients.
We first applied liver transcriptomics41 to identify differentially expressed mRNAs as candidate biomarkers for the early detection of HBV-related liver injuries. STEM and WGCNA analyses were employed in the next steps, which can accurately group distinct expression patterns42 and identify numerous intra-modular hub mRNAs,43,44 respectively. A total of 9 progressively elevated mRNAs were identified as the hub mRNAs by STEM and WGCNA (Figure 2). The expression trends of the selected mRNAs were verified by qRT-PCR and immunohistochemistry, which indicated that SHC1, SLAMF8 and IL-32 levels were in line with the sequencing results.
SHC1 belongs to a family of adapter proteins that contain p66Shc, p52Shc and p46Shc isoforms.20,45 Among the three isoforms, p66Shc has an additional CH2 domain containing a S36 residue, which attributes to its dual functionality in oxidative stress, cell growth, proliferation and apoptosis. Mir et al20 reported that p66Shc might act as a valuable biomarker for the prognosis of metabolic disorders, chronic age-related diseases and cancer by regulating ROS levels. Pei et al46 showed a physiological function for p66Shc in tuning the internal physiology of an organism to the external environment by modulating the rhythm of endogenous H2O2 levels. Interestingly, all these isoforms have been proven to be upregulated in LF18,47,48 and HCC.16,19,49 As consistent with these studies, our results demonstrated that SHC1 levels in HCC and LF/LC patients were higher than those in CHB patients and healthy individuals. Previously, Yoshida et al17 found that p46shc was overexpressed in the cytoplasm and nucleus of tumor cells and/or hepatocytes in HCC rats, whereas p52shc was localized in the cytoplasm. In addition, Huang et al16 showed that p66Shc staining was enriched in the cytoplasm. Interestingly, we found that SHC1 was mainly located in the Kupffer cell cytoplasm, and to a lesser extent, in the hepatocellular cytoplasm (Figure 4). SLAMF8/CD353 is a member of the SLAMF receptor family that modulates the activation and functions of many immune cells.50,51 Previous studies indicated that SLAMF8 was a diagnostic and prognostic marker for pulmonary tuberculosis,31 glioma23 and anaplastic large cell lymphoma.51 Zeng et al25 demonstrated that the combined deficiency of SLAMF8 and SLAMF9 could prevent endotoxin-induced hepatic inflammation, and could act as the promising therapeutic targets for acute liver injury. However, our findings demonstrated a significant increase in the expression of SLAMF8 in HBV-related liver injuries, and its level increased with the severity of these injuries (Figures 3 and 4). IL-32 is a pro-inflammatory cytokine that has been associated with a variety of inflammatory disorders and tumors.28,52 Previous research has demonstrated that HBV can induce the expression of IL-32.53 IL-32 is also upregulated in HBV-infected liver inflammation,28 fibrosis4 and carcinoma.29 In our study, compared with healthy controls, patients with severe HBV-related liver diseases exhibited significantly higher levels of IL-32. As IL-32 protein is expressed in almost all hepatocytes and localized in the cytoplasm,29 our IHC staining revealed that IL-32 was found predominantly in the cytoplasm of HBV-associated liver tissues (Figure 4).
Sensitive and specific biomarkers are essential for the early detection and diagnosis of hepatitis, hepatic fibrosis and HCC, as well as the development of preventive screening.54 Many researches have been carried out to develop biomarkers, but only a few of them have been translated from the research laboratory into the routine clinical practice.31,55 Considering that plasma-based biomarkers can facilitate the clinical diagnosis of diseases,56 we established a comprehensive mRNA-based model that could be implicated in HCC, LF/LC and CHB. It was found that the plasma levels of 3 identified mRNAs, such as SHC1, SLAMF8 and IL-32, were gradually elevated during the progression of HBV-associated liver diseases (Table 2; Supplemental Table 2). Furthermore, by combining the 3-mRNA panel and 3 clinical parameters, the diagnosis accuracy for differentiating between “HCC and LF/LC”, “LF/LC and CHB”, and “CHB and healthy controls” could be markedly improved (Figure 5; Figure S2).
Nevertheless, there are some inevitable limitations that need to be addressed. Firstly, the present study was conducted at only one hospital and the findings should be verified in large, multicenter, prospective cohort studies.55 Secondly, our study was solely focused on Chinese population with HBV-associated liver diseases. Thus, future research is needed to confirm the generalizability of our results by covering other ethnicities, geographical areas, and risk factors, including alcohol abuse, hepatitis C virus infection, etc.5 Thirdly, although SHC1, SLAMF8 and IL-32 were stably expressed in plasma, their secretion and transport to the circulation remain largely unclear. Therefore, additional functional studies are warranted to provide a new insight into the molecular mechanisms of the identified mRNAs in HBV-related disease pathogenesis.
Conclusions
In summary, transcriptome sequencing and bioinformatics analysis were performed to establish a panel of hub mRNAs for HBV-associated liver diseases. Notably, SHC1, SLAMF8, IL-32 and APFSSI model could differentiate between “HCC patients and LF/LC patients”, “LF/LC patients and CHB patients”, and “CHB patients and healthy controls”. The performance of the APFSSI model for discriminating CHB from healthy subjects (AUC=0.966) was much greater compared to SHC1 (AUC=0.900), SLAMF8 (AUC=0.744) and IL-32 (AUC=0.821). When distinguishing LF/LC from CHB, APFSSI was the most outstanding diagnostic parameter (AUC=0.924), which was superior to SHC1, SLAMF8 and IL-32 (AUC=0.812, 0.684 and 0.741, respectively). The differentiation power of the APFSSI model for HCC vs LF/LC (AUC=0.904) was far superior to SHC1 (AUC=0.646), SLAMF8 (AUC=0.802) and IL-32 (AUC=0.761). Using the APFSSI values of 1.624, 2.470 and 3.349 as the cut-off point, we could accurately diagnose CHB, LF/LC and HCC, respectively. To our knowledge, this study is the first to provide threshold points for the selected mRNAs and APFSSI model, which may accelerate their bench to bedside translation and improve the management of HBV-related liver injuries.
Abbreviations
AFP, alpha fetoprotein; ALB, albumin; ALT, alanine aminotransferase; AST, aspartate aminotransferase; APRI, AST-to-PLT ratio index; AUC, area under the curve; CHB, chronic hepatitis B; CI, confidence interval; CREA, creatinine; EFEMP1, EGF containing fibulin extracellular matrix protein 1; ELISA, enzyme-linked immunosorbent assay; FIB-4, fibrosis index based on the 4 factor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GGT, gamma-glutamyl transferase; GO, Gene Ontology; HBV, hepatitis B virus; HBsAg, hepatitis B surface antigen; HCC, hepatocellular carcinoma; HGB, hemoglobin; IHC, Immunohistochemical staining; INR, international normalized ratio; IL-32, interleukin-32; IOD, integral optical density; ITGB2, integrin subunit beta 2; LF, liver fibrosis; LC, liver cirrhosis; LSM, liver stiffness measurement; MANF, mesencephalic astrocyte derived neurotrophic factor; PBMC, peripheral blood mononuclear cell; PLT, platelet; PT, prothrombin time; qRT-PCR, quantitative reverse-transcriptase polymerase chain reaction; ROC, receiver operating characteristic; SHC1, SHC adaptor protein 1; SLAMF8, SLAM family member 8; STEM, short time-series expression miner; SYNPO, synaptopodin; TC2N, tandem C2 domains, nuclear; TBIL, total bilirubin; TOM, topological overlap matrix; UGP2, UDP-glucose pyrophosphorylase 2; WBC, white blood cell; WGCNA, weighted gene co-expression network analysis.
Data Sharing Statement
The authors declare that all relevant data supporting the findings presented in this study are available within the article and its Supplementary Information files, or, from the corresponding author upon reasonable request.
Ethics Approval and Informed Consent
This study was approved by the local ethics committee of the Third Hospital of Hebei Medical University (K2019-014-2), and a written informed consent was also obtained from each participant.
Acknowledgments
The authors would like to express their gratitude to Liang Qiao for the guidance of case selection.
Author Contributions
Concept, design, and writing of the article: Dandan Zhao; execution: Xiaoxiao Zhang, Yuhui Tang, Peilin Guo, Rong Ai; acquisition of data: Mengmeng Hou, Yiqi Wang, Xiwei Yuan, Luyao Cui; analysis and interpretation: Yuguo Zhang, Suxian Zhao, Wencong Li, Yang Wang, Xiaoye Sun, Lingdi Liu, Shiming Dong, Lu Li, Wen Zhao; critical revision of the article and financial support: Yuemin Nan. All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
The study was supported by the Key Research and Development Program of Hebei Province, No. 19277779D, The Program of Introduce International Intelligence of Hebei Province, and The Forth Batch of High-talents in Hebei Province.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sun Z, Liu X, Wu D, et al. Circulating proteomic panels for diagnosis and risk stratification of acute-on-chronic liver failure in patients with viral hepatitis B. Theranostics. 2019;9(4):1200–1214. doi:10.7150/thno.31991
2. Wang X, Li MM, Niu Y, et al. Serum zonulin in HBV-associated chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Dis Markers. 2019;2019:5945721. doi:10.1155/2019/5945721
3. Li L-J, Wu X-Y, Tan S-W, et al. Lnc-TCL6 is a potential biomarker for early diagnosis and grade in liver-cirrhosis patients. Gastroenterol Rep. 2019;7(6):434–443. doi:10.1093/gastro/goz050
4. Xu H, Zhang S, Pan X, et al. TIMP-1 expression induced by IL-32 is mediated through activation of AP-1 signal pathway. Int Immunopharmacol. 2016;38:233–237. doi:10.1016/j.intimp.2016.06.002
5. Cai J, Chen L, Zhang Z, et al. Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut. 2019;68(12):2195–2205. doi:10.1136/gutjnl-2019-318882
6. Tseng TC, Liu CJ, Su TH, et al. Fibrosis-4 index helps identify HBV carriers with the lowest risk of hepatocellular carcinoma. Am J Gastroenterol. 2017;112(10):1564–1574. doi:10.1038/ajg.2017.254
7. Ji D, Chen GF, Wang JC, et al. Identification of TAF1, HNF4A, and CALM2 as potential therapeutic target genes for liver fibrosis. J Cell Physiol. 2018;234(6):9045–9051. doi:10.1002/jcp.27579
8. Xie G, Wang X, Wei R, et al. Serum metabolite profiles are associated with the presence of advanced liver fibrosis in Chinese patients with chronic hepatitis B viral infection. BMC Med. 2020;18(1):144. doi:10.1186/s12916-020-01595-w
9. Kobayashi K, Nakao H, Nishiyama T, et al. Diagnostic accuracy of real-time tissue elastography for the staging of liver fibrosis: a meta-analysis. Eur Radiol. 2015;25(1):230–238. doi:10.1007/s00330-014-3364-x
10. Kang YH, Park MY, Yoon DY, et al. Dysregulation of overexpressed IL-32 alpha in hepatocellular carcinoma suppresses cell growth and induces apoptosis through inactivation of NF-kappaB and Bcl-2. Cancer Lett. 2012;318(2):226–233. doi:10.1016/j.canlet.2011.12.023
11. Qian X, Liu S, Long H, et al. Reappraisal of the diagnostic value of alpha‐fetoprotein for surveillance of HBV‐related hepatocellular carcinoma in the era of antiviral therapy. J Viral Hepat. 2020;28(1):20–29. doi:10.1111/jvh.13388
12. Papatheodoridis G, Dalekos G, Sypsa V, et al. PAGE-B predicts the risk of developing hepatocellular carcinoma in Caucasians with chronic hepatitis B on 5-year antiviral therapy. J Hepatol. 2016;64(4):800–806. doi:10.1016/j.jhep.2015.11.035
13. Sharma SA, Kowgier M, Hansen BE, et al. Toronto HCC risk index: a validated scoring system to predict 10-year risk of HCC in patients with cirrhosis. J Hepatol. 2017. doi:10.1016/j.jhep.2017.07.033
14. Fan R, Papatheodoridis G, Sun J, et al. aMAP risk score predicts hepatocellular carcinoma development in patients with chronic hepatitis. J Hepatol. 2020;73(6):1368–1378. doi:10.1016/j.jhep.2020.07.025
15. Xu RH, Wei W, Krawczyk M, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater. 2017;16(11):1155–1161. doi:10.1038/nmat4997
16. Huang P, Feng X, Zhao Z, et al. p66Shc promotes HCC progression in the tumor microenvironment via STAT3 signaling. Exp Cell Res. 2019;383(2):111550. doi:10.1016/j.yexcr.2019.111550
17. Yoshida S, Masaki T, Feng H, et al. Enhanced expression of adaptor molecule p46 Shc in nuclei of hepatocellular carcinoma cells: study of LEC rats. Int J Oncol. 2004;25(4):1089–1096.
18. Ma H, Wang C, Liu X, Zhan M, Wei W, Niu J. Src homolog and collagen homolog1 isoforms in acute and chronic liver injuries. Life Sci. 2021;273:119302. doi:10.1016/j.lfs.2021.119302
19. Zhang R, Lin XH, Ma M, et al. Periostin involved in the activated hepatic stellate cells-induced progression of residual hepatocellular carcinoma after sublethal heat treatment: its role and potential for therapeutic inhibition. J Transl Med. 2018;16(1):302. doi:10.1186/s12967-018-1676-3
20. Mir HA, Ali R, Mushtaq U, Khanday FA. Structure-functional implications of longevity protein p66Shc in health and disease. Ageing Res Rev. 2020;63:101139. doi:10.1016/j.arr.2020.101139
21. Li X, Kang C, Geng B, et al. PTRF/CAVIN1, regulated by SHC1 through the EGFR pathway, is found in urine exosomes as a potential biomarker of ccRCC. Carcinogenesis. 2020;41(3):274–283. doi:10.1093/carcin/bgz147
22. Qin W, Rong X, Yu C, Jia P, Yang J, Zhou G. Knockout of SLAMF8 attenuates collagen-induced rheumatoid arthritis in mice through inhibiting TLR4/NF-kappaB signaling pathway. Int Immunopharmacol. 2022;107:108644. doi:10.1016/j.intimp.2022.108644
23. Zou CY, Guan GF, Zhu C, et al. Costimulatory checkpoint SLAMF8 is an independent prognosis factor in glioma. CNS Neurosci Ther. 2018;25(3):333–342. doi:10.1111/cns.13041
24. Zhang Q, Cheng L, Qin Y, et al. SLAMF8 expression predicts the efficacy of anti-PD1 immunotherapy in gastrointestinal cancers. Clin Transl Immunol. 2021;10(10):e1347. doi:10.1002/cti2.1347
25. Zeng X, Liu G, Peng W, et al. Combined deficiency of SLAMF8 and SLAMF9 prevents endotoxin-induced liver inflammation by downregulating TLR4 expression on macrophages. Cell Mol Immunol. 2020;17(2):153–162. doi:10.1038/s41423-018-0191-z
26. de Albuquerque R, Komsi E, Starskaia I, Ullah U, Lahesmaa R. The role of Interleukin-32 in autoimmunity. Scand J Immunol. 2021;93(2):e13012. doi:10.1111/sji.13012
27. Sloot YJE, Smit JW, Joosten LAB, Netea-Maier RT. Insights into the role of IL-32 in cancer. Semin Immunol. 2018;38:24–32. doi:10.1016/j.smim.2018.03.004
28. Kim DH, Park ES, Lee AR, et al. Intracellular interleukin-32 gamma mediates antiviral activity of cytokines against hepatitis B virus. Nat Commun. 2018;9(1):3284. doi:10.1038/s41467-018-05782-5
29. Zhao WB, Wang QL, Xu YT, Xu SF, Qiu Y, Zhu F. Overexpression of interleukin-32α promotes invasion by modulating VEGF in hepatocellular carcinoma. Oncol Rep. 2018;39(3):1155–1162. doi:10.3892/or.2017.6162
30. Wu D, Zhang S, Xie Z, et al. Plasminogen as a prognostic biomarker for HBV-related acute-on-chronic liver failure. J Clin Invest. 2020;130(4):2069–2080. doi:10.1172/JCI130197
31. Zhao G, Luo X, Han X, Liu Z. Combining bioinformatics and biological detection to identify novel biomarkers for diagnosis and prognosis of pulmonary tuberculosis. Saudi Med J. 2020;41(4):351–360. doi:10.15537/smj.2020.4.24989
32. Lampertico P, Agarwal K, Berg T; European Association for the Study of the Liver. Electronic address eee, European Association for the Study of the L. EASL 2017 clinical practice guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67(2):370–398. doi:10.1016/j.jhep.2017.03.021
33. Terrault NA, Lok ASF, McMahon BJ, et al. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology. 2018;67(4):1560–1599. doi:10.1002/hep.29800
34. Vogel A, Cervantes A, Chau I, et al. Hepatocellular carcinoma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29(Suppl 4):iv238–iv255. doi:10.1093/annonc/mdy308
35. Li D, Zhao D, Du J, et al. Heme oxygenase-1 alleviated non-alcoholic fatty liver disease via suppressing ROS-dependent endoplasmic reticulum stress. Life Sci. 2020;253:117678. doi:10.1016/j.lfs.2020.117678
36. Xu H, Zhang A, Han X, et al. ITGB2 as a prognostic indicator and a predictive marker for immunotherapy in gliomas. Cancer Immunol Immunother. 2022;71(3):645–660. doi:10.1007/s00262-021-03022-2
37. Sun Z, Dang Q, Liu Z, et al. LINC01272/miR-876/ITGB2 axis facilitates the metastasis of colorectal cancer via epithelial-mesenchymal transition. J Cancer. 2021;12(13):3909–3919. doi:10.7150/jca.55666
38. Zhang L, Huang Y, Ling J, et al. Is integrin subunit alpha 2 expression a prognostic factor for liver carcinoma? A validation experiment based on bioinformatics analysis. Pathol Oncol Res. 2019;25(4):1545–1552. doi:10.1007/s12253-018-0551-0
39. He J, Li G, Liu X, et al. Diagnostic and prognostic values of MANF expression in hepatocellular carcinoma. Biomed Res Int. 2020;2020:1936385. doi:10.1155/2020/1936385
40. Liu J, Wu Z, Han D, et al. Mesencephalic astrocyte-derived neurotrophic factor inhibits liver cancer through Small Ubiquitin-Related Modifier (SUMO)ylation-related suppression of NF-kappaB/snail signaling pathway and epithelial-mesenchymal transition. Hepatology. 2020;71(4):1262–1278. doi:10.1002/hep.30917
41. Baselli GA, Dongiovanni P, Rametta R, et al. Liver transcriptomics highlights interleukin-32 as novel NAFLD-related cytokine and candidate biomarker. Gut. 2020;69(10):1855–1866. doi:10.1136/gutjnl-2019-319226
42. Qi F, Wang L, Huang P, Zhao Z, Yang B, Xia J. Time-series clustering of cytokine expression after transarterial chemoembolization in patients with hepatocellular carcinoma. Oncol Lett. 2020;19(2):1175–1186. doi:10.3892/ol.2019.11209
43. Liu H, Liu M, You H, Li X, Li X. Oncogenic network and hub genes for natural killer/T-cell lymphoma utilizing WGCNA. Front Oncol. 2020;10:223. doi:10.3389/fonc.2020.0022
44. Hendrickx DM, Jennen DG, Briedé JJ, Cavill R, de Kok TM, Kleinjans JC. Pattern recognition methods to relate time profiles of gene expression with phenotypic data: a comparative study. Bioinformatics. 2015;31(13):2115–2122. doi:10.1093/bioinformatics/btv108
45. Grossman SR, Lyle S, Resnick MB, et al. p66 Shc tumor levels show a strong prognostic correlation with disease outcome in stage IIA colon cancer. Clin Cancer Res. 2007;13(19):5798–5804. doi:10.1158/1078-0432.CCR-07-0073
46. Pei JF, Li XK, Li WQ, et al. Diurnal oscillations of endogenous H2O2 sustained by p66(Shc) regulate circadian clocks. Nat Cell Biol. 2019;21(12):1553–1564. doi:10.1038/s41556-019-0420-4
47. Zhao Y, Wang Z, Zhou J, et al. LncRNA Mical2/miR-203a-3p sponge participates in epithelial-mesenchymal transition by targeting p66Shc in liver fibrosis. Toxicol Appl Pharmacol. 2020;403:115125. doi:10.1016/j.taap.2020.115125
48. Zhao Y, Wang Z, Feng D, et al. p66Shc contributes to liver fibrosis through the regulation of mitochondrial reactive oxygen species. Theranostics. 2019;9(5):1510–1522. doi:10.7150/thno.29620
49. Yoshida S, Kornek M, Ikenaga N, et al. Sublethal heat treatment promotes epithelial-mesenchymal transition and enhances the malignant potential of hepatocellular carcinoma. Hepatology. 2013;58(5):1667–1680. doi:10.1002/hep.26526
50. De Calisto J, Wang N, Wang G, Yigit B, Engel P, Terhorst C. SAP-dependent and -independent regulation of innate T cell development involving SLAMF receptors. Front Immunol. 2014;5:186. doi:10.3389/fimmu.2014.00186
51. Sugimoto A, Kataoka TR, Ito H, et al. SLAM family member 8 is expressed in and enhances the growth of anaplastic large cell lymphoma. Sci Rep. 2020;10(1):2505. doi:10.1038/s41598-020-59530-1
52. Hong JT, Son DJ, Lee CK, Yoon DY, Lee DH, Park MH. Interleukin 32, inflammation and cancer. Pharmacol Ther. 2017;174:127–137. doi:10.1016/j.pharmthera.2017.02.025
53. Tian ZJ, Shen Y, Li XR, Wei YN, Fan H, Ren QK. Increased interleukin-32, interleukin-1, and interferon-gamma levels in serum from hepatitis B patients and in HBV-stimulated peripheral blood mononuclear cells from healthy volunteers. J Infect Public Health. 2019;12(1):7–12. doi:10.1016/j.jiph.2018.06.006
54. Zhu HT, Liu RB, Liang YY, et al. Serum microRNA profiles as diagnostic biomarkers for HBV-positive hepatocellular carcinoma. Liver Int. 2017;37(6):888–896. doi:10.1111/liv.13356
55. Gao S, Xu X, Wang Y, Zhang W, Wang X. Diagnostic utility of plasma lncRNA small nucleolar RNA host gene 1 in patients with hepatocellular carcinoma. Mol Med Rep. 2018;18(3):3305–3313. doi:10.3892/mmr.2018.9336
56. Wu D, Zhang P, Ma J, et al. Serum biomarker panels for the diagnosis of gastric cancer. Cancer Med. 2019;8(4):1576–1583. doi:10.1002/cam4.2055
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.