Back to Journals » Journal of Blood Medicine » Volume 12
Identification and Validation of Hemophilia-Related Outcomes on Japanese Electronic Medical Record Database (Hemophilia-REAL V Study)
Authors Fujiwara T , Miyakoshi C, Kanemitsu T, Okumura Y , Tokumasu H
Received 4 April 2021
Accepted for publication 22 June 2021
Published 6 July 2021 Volume 2021:12 Pages 571—580
DOI https://doi.org/10.2147/JBM.S313371
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Takashi Fujiwara,1,2 Chisato Miyakoshi,3 Takashi Kanemitsu,4 Yasuyuki Okumura,5 Hironobu Tokumasu2,6
1Department of Management, Clinical Research Center, Kurashiki Central Hospital, Okayama, Japan; 2Department of Public Health Research, Kurashiki Clinical Research Institute, Okayama, Japan; 3Department of Pediatrics, Kobe City Medical Center General Hospital, Hyogo, Japan; 4Medical Affairs Division, Chugai Pharmaceutical Co., Ltd, Tokyo, Japan; 5Department of Psychiatry and Behavioral Science, Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan; 6Real World Data Co., Kyoto, Japan
Correspondence: Takashi Fujiwara
Department of Management, Clinical Research Center, Kurashiki Central Hospital, 1-1-1 Miwa, Kurashiki City, Okayama, Japan
Tel +81-86-422-0210
Fax +81-86-421-3424
Email [email protected]
Purpose: Routinely collected data are useful for epidemiological study in hemophilia, but few studies validated the algorithm accuracy. We aimed to develop and validate algorithms to identify patients with hemophilia A and hemophilia A-related events.
Patients and Methods: This validation study compared data from medical chart reviews to a database of routinely collected health data, including claims data and discharge abstracts, and especially electronic medical records (EMR), at a single Japanese hospital (Kurashiki Central Hospital) using a stratified sampling method. Two physicians reviewed the charts for all patients at high risk for hemophilia A, and randomly sampled patients with moderate risk. Diagnostic accuracy was determined based on sensitivity, specificity, positive predictive value (PPV), and negative predictive value.
Results: There were 1,033,845 eligible patients, of whom 31 had a diagnosis of hemophilia A. ICD-10 diagnosis code D66 in the EMR identified hemophilia A with a sensitivity of 93.5% (95% confidence interval: 78.6– 99) and PPV of 61.7% (95% confidence interval: 46.4– 75.5). The administration of ≥ 10,000 units/month of factor VIII products, as documented in the EMR, identified 81.3% of patients with prophylactic factor replacement therapy. The ICD-10 diagnosis code for intracranial bleeding in the EMR identified 75.0% of patients with intracranial bleeding, but those of gastrointestinal bleeding and major joint bleeding identified only 11.1% and 1.7%, respectively.
Conclusion: We developed and validated algorithms to identify congenital hemophilia A and hemophilia A-related events. Hemophilia A could be identified with high sensitivity and PPV, but it was still challenging to identify hemophilia A-related events.
Keywords: congenital hemophilia, electronic health record, positive predictive value, sensitivity, validation study
Introduction
Congenital hemophilia A is a rare, chronic, heritable bleeding disorder caused by deficiency of clotting factor VIII.1 Hemophilia A is the most common type of congenital hemophilia, occurring in approximately 1 in 5000 live-born males.2 Severe hemophilia A is defined as factor VIII activity of <1%, as seen in two-thirds of hemophilia A patients. Patients with hemophilia A suffer from lifelong bleeding, but the availability of factor replacement products has markedly improved the care for patients with these conditions over the past decade.3,4
The low prevalence of hemophilia A complicates large-scale epidemiological studies. Therefore, routinely collected health data, such as electronic medical records (EMR) and claims data, are vital to understand the clinical course of hemophilia A.5,6 Accurate identification of patients with hemophilia A is crucial, but misclassification can occur. Algorithms for identifying patients with hemophilia A in EMR and claims databases have been developed and validated in the United States.7,8 However, there have been no reports of validation studies of hemophilia in Japan. Health care systems, including EMR and claims data, vary among countries; therefore, the accuracy of algorithms for identifying disease may also vary. It is important to develop algorithms for identifying patients with hemophilia A to conduct epidemiological studies using EMR and claims data. In addition, no study has developed a validated algorithm to identify hemophilia A-related outcomes.7,8 The present study was performed to develop and validate algorithms for identifying patients with hemophilia A and hemophilia A-related outcomes using the EMR and claims database in Japan.
Patients and Methods
Study Design
This validation study compared data from medical chart reviews to a database of routinely collected health data, including EMR, claims data, and discharge abstracts for a single Japanese hospital (Kurashiki Central Hospital) using a stratified sampling method.9 Kurashiki Central Hospital is an urban hospital with 1172 beds that serves 800,000 people in the western area of Okayama Prefecture, Japan.10
This study was conducted and reported in accordance with the statement of the Japanese Society for Pharmacoepidemiology and the Standards for Reporting of Diagnostic Accuracy Studies criteria.11–13 The study was approved by the institutional review board of Kurashiki Central Hospital and the Research Institute of Healthcare Data Science. This study was registered in the UMIN Clinical Trials Registry (Trial Number: UMIN000038212; https://www.umin.ac.jp/ctr/index-j.htm).
Data Sources
This study used anonymized, routinely collected health data stored in a database.10 The Health, Clinic, and Education Information Evaluation Institute (HCEI) has contracts with more than 190 healthcare institutions, including Kurashiki Central Hospital, to collect EMR and claims data from those institutions and develop a large-scale database, known as the RWD database. The anonymized data for this research study were collected by the HCEI on August 28, 2019. The RWD database included approximately 20.5 million inpatients and outpatients. In the database, disease data are extracted from EMRs and are recorded based on the International Classification of Diseases, 10th revision (ICD-10) codes. Drugs are labeled based on the Japanese receipt code and YJ code. Laboratory test results are standardized and labeled according to the Japanese Laboratory Code version 10.
Study Population
Due to the low prevalence of hemophilia A, it would be difficult to ensure precise diagnostic value with a random sample of patients from the overall patients at Kurashiki Central Hospital. Therefore, we used a stratified sampling method to identify all possible cases of hemophilia A.11,14,15
Patients were classified as a having high, moderate, or low risk for hemophilia A based on diagnostic codes (ICD-10), drug codes, procedural codes, and notes in the medical records. Patients who received an ICD-10 diagnosis or suspected code D66 (congenital factor VIII disorder, hemophilia A) were classified as being at high risk for hemophilia A. We defined moderate risk of hemophilia A as follows: diagnosed or suspected congenital factor IX disorder or hemophilia B (ICD-10 D67), von Willebrand disease (D680), or acquired factor VIII disorder or hemophilia A (D684); prescription for hemophilia treatment (factor VIII products); blood test related to hemophilia (factor VIII activity); and “hemophilia A” in chart notes (see Appendix 1). We defined all other patients as being at low risk for hemophilia A. Chart review was conducted for all high-risk patients and randomly sampled moderate-risk patients.
Ascertaining Hemophilia A and Disease-Related Outcomes
Detailed medical chart reviews of all high-risk patients and a subset of moderate-risk patients were conducted. Hemophilia A-related outcomes including disease, treatment and disease-related events were analyzed. Two physicians (an adult hematologist and a pediatric hematologist) independently conducted the paper and electrical chart reviews for all available periods. Kurashiki Central Hospital introduced an EMR system in 2003, before which paper charts were used. The reviewers also identified hemophilia A-related outcomes, including disease characteristics (severity and history of factor VIII inhibitor), treatment issue (prophylactic factor VIII replacement therapy), and disease-related events (intracranial bleeding, gastrointestinal bleeding, and major joint bleeding). Disagreements were resolved by discussion.
Data Linkage and Data Extraction from Database
The RWD database contained healthcare EMR data from individual medical institutions. When extracting EMR data from medical institutions, patients’ EMR numbers were removed, and each patient was hash-encoded to anonymize the records. We also used chart review data, which contained personal identification information; these data were anonymized, and a hash was provided encoding the RWD for each patient. The chart review data were subsequently linked with the RWD database.
Statistical Analysis
The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of each algorithm were calculated, accepting the chart review results as the reference standard. When evaluating the diagnostic values of disease (hemophilia A), we calculated these diagnostic values adjusted with weighted sampling according to a task force report on the validation of diagnosis codes in Japan.11 When calculating the diagnostic values of disease characteristics (severity and history of factor VIII inhibitor) and treatment issue (prophylactic factor VIII replacement therapy), we examined data for patients with ICD-10 diagnosis or suspected code D66. When assessing disease-related events (intracranial bleeding, gastrointestinal bleeding, and major joint bleeding), we determined the sensitivity and PPV. If a disease-related event occurred within 3 days of the recorded outcome, it was recorded as a true positive.
For evaluating the diagnostic value of severity of hemophilia A, the algorithm used laboratory test results of factor VIII activity. Laboratory test results of factor VIII activity are often stored as numeric data in the RWD database, but the results are sometimes stored as characteristic data, which we defined as follows: severe; ≤1, <2, and <3, moderate; ≤5; mild; <5.0, <5.1, and ≥5.0, normal; (+).
In accordance with the task force report on the validation of diagnosis codes in Japan,11 we evaluated how representative the population in this validation study was of the entire RWD population. We compared patients who received an ICD-10 diagnosis code D66 in Kurashiki Central Hospital and those in the RWD database. We assessed patient characteristics (age at data extraction, sex), comorbidities (hypertension, diabetes mellitus, hyperlipidemia, arteriosclerosis, and cardiovascular disease), and hemophilia-related data (age at receiving ICD-10 diagnosis code D66, factor VIII activity, factor VIII inhibitor, prescription of factor VIII products, and prophylactic factor replacement therapy). The presence of comorbidities was defined using the ICD-10 diagnosis codes (Appendix 2).
Statistical analyses were performed using R software (version 3.4.1; R Foundation for Statistical Computing, Vienna, Austria).
Study Ethics
This study was approved by the institutional ethics committee of the Research Institute of Healthcare Data Science (https://rihds.org/ethic/) and the institutional ethics committee of Kurashiki Central Hospital. Optout consent was used.
Results
Patient Selection
We identified 128 patients with a high risk of hemophilia A from the medical records of Kurashiki Central Hospital. We also identified 895 patients with moderate risk of hemophilia A and randomly selected 120 of these patients (Figure 1). We conducted a chart review of the 248 patients, and the data were linked with the RWD database. The chart review revealed no hemophilia patients among those with a moderate risk of the disease. After data linkage of the chart review and the RWD database, 12 patients were excluded due to a lack of data in the RWD database (Appendix 3).
 |
Figure 1 Flow diagram of patients’ selection and validation. Note: *We used Japanese written language (Kanji) when searching of “hemophilia A” in electronic medical record (EMR). |
Of the 236 patients included in the study, 31 were identified as hemophilia A and were in the high-risk group. There were no cases of hemophilia A in the moderate-risk group (Figure 1). The characteristics of the 31 patients with hemophilia A are shown in Table 1.
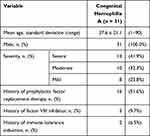 |
Table 1 Characteristics of Patients with Congenital Hemophilia A of Kurashiki Central Hospital |
Diagnostic Value
The definitions of each algorithm and the diagnostic values are shown in Tables 2–4. In the outcome condition of disease (hemophilia A), ICD-10 diagnosis or suspected code D66 showed 100% sensitivity, with low PPV (24.4%). Compared with ICD-10 diagnosis or suspected code D66, ICD-10 diagnosis code D66, and male sex had similar sensitivity (93.5%) but higher PPV (73.3%; Table 2). Among 47 patients with a definitive diagnosis of hemophilia (ICD-10 D66) according to the Kurashiki Central Hospital EMR database, the sensitivity and specificity of disease characteristics and treatment issues were about 70–90% (Table 3). However, the sensitivity and PPV of disease-related events were very low except for intracranial bleeding (Table 4). The data for other diagnoses are listed in Appendix 4. The definition of treatment products are listed in Appendix 5, Appendix 6, and Appendix 7.
 |
Table 2 Diagnostic Value of Algorithm of Outcome Conditions: Disease (Hemophilia A) |
 |
Table 3 Diagnostic Value of Algorithm of Outcome Conditions: Disease Characteristics and Treatment |
 |
Table 4 Diagnostic Value of Algorithms of Outcome Conditions: Hemophilia A-Related Events |
Comparison of Kurashiki Central Hospital Data and the RWD Database
In the outcome definition of hemophilia A, the ICD10 diagnosis code D66 algorithm using EMR data had useful diagnostic value. Therefore, we used this algorithm to assess how representative the population in this study was of the entire RWD database. Table 5 shows the patients receiving ICD-10 diagnosis code D66 in Kurashiki Central Hospital and the RWD database. The definition of co-morbidity disease for the comparison are listed in Appendix 8.
 |
Table 5 Characteristics of Patients with a Diagnosis of ICD-10 D66 |
Discussion
Japan has recently promoted the use of RWD, including EMR and claims databases,16 but there have been no comprehensive validation studies. A recent systematic review suggested that only six validation studies had been performed in Japan as of 2017.17 Several studies identified and validated algorithms of hemophilia A.7,8 However, these studies only validated the algorithms of disease condition and not algorithms of hemophilia A-related events (eg, joint bleeding, and intracranial bleeding). By contrast, the present study revealed the diagnostic value of disease condition, treatment condition, and hemophilia A-related events. The results of this study will promote epidemiological studies of hemophilia A using the EMR and claims database.
Algorithm for Hemophilia A
The sensitivity of the ICD-10 diagnosis or suspected code D66 algorithm was 100.0%, but the PPV was 24.4%. The sensitivity of the ICD-10 diagnosis code D66 algorithm had a higher PPV (61.7%), but the sensitivity was 93.5%. Of the 31 hemophilia patients identified in this study, the treatment period in Kurashiki Central Hospital varied. Two patients were treated 30 years ago, and the medical records were mainly stored in paper charts; these patients appeared as false negatives in the ICD-10 diagnosis code D66 algorithm. The sensitivity and PPV would be higher when evaluating diagnostic values using populations currently undergoing treatment.
Algorithm for Disease-Related Events
This study identified and validated an algorithm for hemophilia A-related events. In hemophilia A, clinical practice guidelines define intracranial, neck/throat, and gastrointestinal bleeding as life-threatening.3 Therefore, we attempted to identify and validate an algorithm for these types of bleeding conditions, although there were no cases of neck/throat bleeding in this study. We also evaluated the diagnostic value of the algorithm for major joint bleeding because that is one of the most frequent hemophilia A-related events.
Intracranial bleeding is the most critical hemophilia A-related event, and the algorithm based on its ICD-10 diagnosis code had 75.0% sensitivity and 33.3% PPV. During the study period, intracranial bleeding occurred in four patients. No disease-related ICD-10 code was provided for one event; the patient had asymptomatic intracranial bleeding after brain tumor resection. Disease-related ICD-10 was provided in the three symptomatic cases. The algorithm based on ICD-10 code for intracranial bleeding identified symptomatic intracranial bleeding with 100% sensitivity.
Strengths of the EMR Database
In the healthcare field, there are two primary types of databases for observational research; claims databases and EMR databases, and previous validation studies of hemophilia A used claims data.8,9 This study used the RWD database, which is classified as an EMR database. Compared with claims databases, EMR databases can provide laboratory test results. In this study, we attempted to identify severe hemophilia A using the test results of factor VIII activity. The minimum test results of factor VIII activity <1% algorithm had a sensitivity of 69.2%and specificity of 76.5%. The algorithm yielded false-negative results in four patients: one of these patients had visited our hospital 30 years ago, and the laboratory test results were not stored; two patients had already been treated when Kurashiki Central Hospital introduced EMR, and factor VIII activity was >1%, and the remaining patient had been treated at another hospital before visiting Kurashiki Central Hospital, and factor VIII activity was >1%. Therefore, the limitations of EMR databases must be considered when conducting epidemiological studies stratified by disease severity. Researchers should exclude patients initially treated ≥10 years ago, and consider the influence of transfer from another hospital.
This study revealed that algorithms based on ICD-10 codes for hemophilia A had at least 90% PPV and NPV. However, the accuracy of algorithms based on the ICD-10 codes for disease-related events was very low. Free-text notes in the EMR were not used in any algorithms because of their unstructured nature. However, use of unstructured free-text notes in algorithms for disease-related events could be a target for future research.
Representativeness of the Validation Dataset
To evaluate how representative the population in this validation study was of the entire population in the RWD database, we compared the characteristics of patients receiving ICD-10 diagnosis code D66 in Kurashiki Central Hospital and the RWD database. The ratio of patients with factor VIII activity level <1% in Kurashiki Central Hospital was high, and patients in Kurashiki Central Hospital tended to have comorbidities. Kurashiki Central Hospital is one of the largest hospitals in Japan and treats cases with greater disease severity and higher rates of complications. It would be useful for physicians to understand these differences in patient characteristics between Kurashiki Central Hospital and the RWD database, which could be explained by the role of Kurashiki Central Hospital.
The population in this study is representative of the entire population (ie, of all patients in the RWD database). Therefore, the sensitivity, specificity, PPV, and NPV data could be applied in other epidemiological studies using the RWD database. However, this validation study was conducted at a single large hospital, so the results may not apply to other hospital settings. Future epidemiological studies using the RWD database should consider performing sensitivity analyses based on the hospital volume data contained therein.
Conclusion
In conclusion, we developed and validated EMR- and claims-based definitions of hemophilia A-related outcomes, including disease, treatment, and disease-related events. These results support outcomes research studies using RWD for hemophilia A.
Acknowledgments
The authors would like to thank Dr. Ueda, Dr. Imai, Ms. Satomi (Clinical Research Coordinator), Ms. Yamaguchi (Clinical Research Coordinator), and Ms. Komatsubara (Clinical Research Coordinator) of Kurashiki Central Hospital (Kurashiki, Japan) for the medical chart review in this study. This study was conducted as a collaboration between Kurashiki Central Hospital (Kurashiki, Japan) and Chugai Pharmaceutical Co., Ltd. (Tokyo, Japan).
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This study was funded by Chugai Pharmaceutical Co., Ltd.
Disclosure
TF has received personal fees and/or grants from Real World Data Co., Ltd and Chugai Pharmaceutical Co., Ltd. CM has received personal fees from Chugai Pharmaceutical Co., Ltd. TK is a full-time employee of Chugai Pharmaceutical Co., Ltd. HT is the Chief Operating Officer at Real World Data Co., and has received personal fees from AYUMI Pharmaceutical Corporation and Chugai Pharmaceutical Co., Ltd. YO has received personal fees from Real World Data Co., Ltd, Cando, Inc., the Japan Medical Data Center, the Japan Medical Research Institute Co., Ltd, Ohara HealthCare Foundation, Merck & Co., Inc., and Otsuka Pharmaceutical Co., Ltd. YO’s present affiliation: Real World Data Co., Ltd. The authors report no other conflicts of interest in this work.
References
1. Bolton-Maggs PH, Pasi KJ. Haemophilias A and B. Lancet. 2003;361(9371):1801–1809. doi:10.1016/S0140-6736(03)13405-8
2. Carcao MD. The diagnosis and management of congenital hemophilia. Semin Thromb Hemost. 2012;38(7):727–734. doi:10.1055/s-0032-1326786
3. Srivastava A, Brewer AK, Mauser-Bunschoten EP, et al. Guidelines for the management of hemophilia. Haemophilia. 2013;19(1):e1–47.
4. Berntorp E, Shapiro AD. Modern haemophilia care. Lancet. 2012;379(9824):1447–1456. doi:10.1016/S0140-6736(11)61139-2
5. National Research Council. The Learning Healthcare System: workshop Summary (IOM Roundtable on Evidence-Based Medicine). Washington, DC: The National Academies Press, 2007.
6. Berger ML, Mamdani M, Atkins D, Johnson ML. Good research practices for comparative effectiveness research: defining, reporting and interpreting nonrandomized studies of treatment effects using secondary data sources: the ISPOR Good Research Practices for Retrospective Database Analysis Task Force Report--Part I. Value Health. 2009;12(8):1044–1052. doi:10.1111/j.1524-4733.2009.00600.x
7. Wang M, Cyhaniuk A, Cooper DL, Iyer NN. Identification of patients with congenital hemophilia in a large electronic health record database. J Blood Med. 2017;8:131–139. doi:10.2147/JBM.S133616
8. Lyons J, Desai V, Xu Y, et al. Development and validation of an algorithm for identifying patients with hemophilia A in an administrative claims database. Value Health. 2018;21(9):1098–1103. doi:10.1016/j.jval.2018.03.008
9. Health, Clinic, and Education Information Evaluation Institute. Secondary use of the RWD database. Available from: https://www.hcei.or.jp/page/application.
10. Fujiwara T, Okamoto H, Ohnishi Y, et al. Diagnostic accuracy of lateral neck radiography in ruling out supraglottitis: a prospective observational study. Emerg Med J. 2015;32(5):348–352. doi:10.1136/emermed-2013-203340
11. Iwagami M, Aoki K, Akazawa M, et al. Validation study using administrative data in Japan Task Force Report. Available from: http://www.jspe.jp/committee/pdf/validationtrr120180523.pdf.
12. Benchimol EI, Manuel DG, To T, Griffiths AM, Rabeneck L, Guttmann A. Development and use of reporting guidelines for assessing the quality of validation studies of health administrative data. J Clin Epidemiol. 2011;64(8):821–829. doi:10.1016/j.jclinepi.2010.10.006
13. Bossuyt PM, Reitsma JB, Bruns DE, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ. 2015;351:h5527. doi:10.1136/bmj.h5527
14. Widdifield J, Ivers NM, Young J, et al. Development and validation of an administrative data algorithm to estimate the disease burden and epidemiology of multiple sclerosis in Ontario, Canada. Mult Scler. 2015;21(8):1045–1054. doi:10.1177/1352458514556303
15. Kumamaru H, Judd SE, Curtis JR, et al. Validity of claims-based stroke algorithms in contemporary Medicare data: reasons for geographic and racial differences in stroke (REGARDS) study linked with medicare claims. Circ Cardiovasc Qual Outcomes. 2014;7(4):611–619. doi:10.1161/CIRCOUTCOMES.113.000743
16. Yamada K, Itoh M, Fujimura Y, et al. The utilization and challenges of Japan’s MID-NET ® medical information database network in postmarketing drug safety assessments: a summary of pilot pharmacoepidemiological studies. Pharmacoepidemiol Drug Saf. 2019;28(5):601–608. doi:10.1002/pds.4777
17. Koram N, Delgado M, Stark JH, et al. Validation studies of claims data in the Asia-Pacific region: a comprehensive review. Pharmacoepidemiol Drug Saf. 2019;28(2):156–170. doi:10.1002/pds.4616
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
