Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 16
Identification and Validation of Autophagy-Related Genes in Chronic Obstructive Pulmonary Disease
Authors Sun S, Shen Y, Wang J, Li J, Cao J, Zhang J
Received 23 October 2020
Accepted for publication 30 December 2020
Published 12 January 2021 Volume 2021:16 Pages 67—78
DOI https://doi.org/10.2147/COPD.S288428
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Dr Richard Russell
Shulei Sun, Yuehao Shen, Jie Wang, Jinna Li, Jie Cao, Jing Zhang
Department of Respiratory and Critical Care Medicine, Tianjin Medical University General Hospital, Tianjin 300052, People’s Republic of China
Correspondence: Jing Zhang
Department of Respiratory and Critical Care Medicine, Tianjin Medical University General Hospital, 154 Anshan Road, Heping District, Tianjin 300052, People’s Republic of China
Tel +86-22-60361612
Fax +86-22-60361720
Email [email protected]
Purpose: Autophagy plays essential roles in the development of COPD. We aim to identify and validate the potential autophagy-related genes of COPD through bioinformatics analysis and experiment validation.
Methods: The mRNA expression profile dataset GSE38974 was obtained from GEO database. The potential differentially expressed autophagy-related genes of COPD were screened by R software. Then, protein–protein interactions (PPI), correlation analysis, gene-ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were applied for the differentially expressed autophagy-related genes. Finally, RNA expression of top five differentially expressed autophagy-related genes was validated in blood samples from COPD patients and healthy controls by qRT-PCR.
Results: A total of 40 differentially expressed autophagy-related genes (14 up-regulated genes and 26 down-regulated genes) were identified between 23 COPD patients and 9 healthy controls. The PPI results demonstrated that these autophagy-related genes interacted with each other. The GO and KEGG enrichment analysis of differentially expressed autophagy-related genes indicated several enriched terms related to autophagy and mitophagy. The results of qRT-PCR showed that the expression levels of HIF1A, CDKN1A, BAG3, ERBB2 and ATG16L1 in COPD patients and healthy controls were consistent with the bioinformatics analysis results from mRNA microarray.
Conclusion: We identified 40 potential autophagy-related genes of COPD through bioinformatics analysis. HIF1A, CDKN1A, BAG3, ERBB2 and ATG16L1 may affect the development of COPD by regulating autophagy. These results may expand our understanding of COPD and might be useful in the treatment of COPD.
Keywords: autophagy, COPD, bioinformatics analysis, gene expression omnibus dataset
Introduction
Chronic obstructive pulmonary disease (COPD) is a chronic respiratory disease characterized by incompletely reversible airway obstruction, with a high mortality and disability rate.1 Compared with healthy individuals, patients with COPD have an increased risk of other diseases, such as lung cancer.2 Previous studies reported that the risk factors of COPD include genetic factors, smoking and airway inflammation.3–5 Accumulating evidence has shown that several biological functions are involved in the pathogenesis of COPD, including cell proliferation, apoptosis and autophagy.6–8 Among these biological functions, autophagy plays essential roles in the development of COPD.
Autophagy is the conserved mechanism that delivers endogenous or exogenous cytoplasmic materials to the lysosomes for degradation.9 Autophagy is related to various diseases including respiratory diseases. For instance, miR-93 regulates tumorigenicity and therapy response of glioblastoma by targeting autophagy.10 In addition, IL-4 induces autophagy in B cells leading to exacerbated asthma.11 Some signaling pathways have been reported to affect the biological function of COPD through autophagy. PI3K/AKT/mTOR pathway regulates autophagy to induce apoptosis of alveolar epithelial cells in COPD.12 However, autophagy-related genes of COPD remain largely unknown and need to be further explored. Exploring and revealing the potential autophagy-related genes of COPD will provide us potential biomarkers to treat COPD.
Ezzie et al completed one COPD-related dataset GSE38974, which analyzed the differentially expressed genes between COPD patients and healthy individuals.13 Their results showed that 70 miRNAs and 2667 mRNAs were differentially expressed in the two groups. In this study, the dataset was analyzed again from other perspectives. We explored the differentially expressed autophagy-related genes of COPD by analyzing the dataset GSE38974 from GEO database. Then, protein–protein interactions (PPI), correlation analysis, gene-ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were applied for the differentially expressed autophagy-related genes. Finally, the expression levels of key differentially expressed genes were further verified in COPD patients and healthy individuals.
Materials and Methods
Autophagy-Related Genes Datasets and Microarray Data
A total of 222 genes were obtained from The Human Autophagy Database (http://www.autophagy.lu/index.html). The mRNA expression profile dataset of GSE38974 was downloaded from GEO (http://www.ncbi.nlm.nih.gov/geo/). GSE38974 is in GPL4133 platform (Agilent-014850 Whole Human Genome Microarray 4x44K G4112F), which included 23 COPD and 9 normal lung tissue samples.
Differentially Expressed Analysis of Autophagy-Related Genes
The normalized expression matrix of microarray data was downloaded from the GSE38974 dataset. Then the probes was annotated with the annotation files from the dataset. The repeatability of data in GSE38974 was verified by principal component analysis (PCA). The “limma” package of R software was used to identify the differentially expressed autophagy-related genes. Genes with an adjusted P-value <0.05 and absolute fold-change value >1.5 were considered as differentially expressed genes. The heatmap, volcano plot and box plot were conducted using “heatmap” and “ggplot2” packages of R software.
PPI Analysis and Correlation Analysis of the Differentially Expressed Autophagy-Related Genes
PPI analysis of differentially expressed autophagy-related genes was analyzed using STRING database (https://string-db.org/) and Cytoscape software (version 3.8.1). The correlation analysis of the differentially expressed autophagy-related genes was identified using Spearman correlation in the “corrplot” package of R software.
GO and KEGG Pathway Enrichment Analysis of Autophagy-Related Genes
GO and KEGG pathway enrichment analysis were conducted in R software using the package “GO plot”. The GO analysis consisted of cellular component (CC), biological process (BP) and molecular function (MF).
COPD Patients and Healthy Individuals
A total of 20 COPD patients (cases) and 20 age-matched healthy individuals (controls) were obtained from the Tianjin Medical University General Hospital between July 2020 and October 2020. The diagnosis of COPD was followed according to the Global Initiative for Chronic Obstructive Pulmonary Disease (GOLD) criteria: patients with post-bronchodilator FEV1/FVC <70%, and <80% predicted FEV1. Patients were excluded if they had bronchial asthma, bronchiectasis, pulmonary fibrosis, lung tumor and tuberculosis. The 20 healthy individuals were recruited from the hospital’s health checkup center. This study was conducted in accordance with the Declaration of Helsinki and approved by the Medical Ethics Committee of the hospital. Written informed consent was obtained from all the participants. Venous blood was collected from all cases and controls who participated in the study.
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
The blood samples from each participant were processed to isolate peripheral blood mononuclear cells (PBMCs) using Ficoll solution (Solarbio Life Sciences, Beijing, China). Total RNA was extracted from PBMCs with RNA Extraction Kit (Omega, Guangzhou, China). Reverse transcription was conducted using PrimeScript RT Master Mix Kit (Takara, Dalian, China). The mRNA level was assessed using TB Green Premix Ex Taq Kit (Takara, Dalian, China) following the instructions. Primers are available in Supplementary Table S1. The relative expression of mRNA was calculated by 2−ΔΔCt method with the normalization to ACTB.
Statistical Analysis
The statistical analyses were performed using R software (version 3.6.2). Gene expression levels of our clinical samples were compared using Student’s t-test. P < 0.05 was considered statistically significant.
Results
Differentially Expressed Autophagy-Related Genes in COPD-Retrospective Analysis of Autophagy-Related Genes
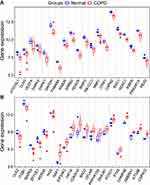
To evaluate the intra-group data repeatability, we performed principal component analysis (PCA), and the results showed that the repeatability of data in GSE38974 is fine (Figure 1A). We next analyzed the expression of 222 autophagy-related genes in 23 COPD patients and 9 healthy individuals, and 40 autophagy-related genes were identified using the criteria of adjusted P-value <0.05 and absolute fold-change value >1.5, including 14 up-regulated genes and 26 down-regulated genes (Table 1). Following the analysis of the GSE38974 dataset with R software, the 40 differentially expressed autophagy-related genes between COPD and normal groups were presented in heatmap and volcano plot (Figure 1B and C). Moreover, box plots showed the expression patterns of 40 differentially expressed autophagy-related genes between COPD and normal samples (Figure 2A and B). The top five up-regulated genes included NAF1, HIF1A, CDKN1A, BAG3 and CHMP4B, and the top five down-regulated genes included ERBB2, GAPDH, FAM48A, ATG16L1, and ITPR1. (Figure 2A and B; Table 1).
 |
Table 1 The 40 Differentially Expressed Autophagy-Related Genes in COPD Samples Compared to Healthy Samples |
PPI Network and Correlation Analysis of the Differentially Expressed Autophagy-Related Genes
To determine the interactions among differentially expressed autophagy-related genes, we performed PPI analysis. The results demonstrated that these autophagy-related genes interacted with each other (Figure 3A) and showed the interaction number of each gene (Figure 3B). To explore the expression correlation of these autophagy-related genes, correlation analysis was performed. The results showed the relationship of the 40 differentially expressed autophagy-related genes in GSE38974 dataset (Figure 4).
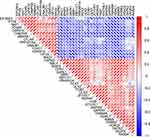 |
Figure 4 Spearman correlation analysis of the 40 differentially expressed autophagy-related genes. |
GO and KEGG Enrichment Analysis of the Differentially Expressed Autophagy-Related Genes
To analyze the potential biological functions of these differentially expressed autophagy-related genes, we conducted GO and KEGG enrichment analysis by using R software. The results revealed that the most significant GO enriched terms involved in autophagy, process utilizing autophagic mechanism, macroautophagy (biological process); autophagosome, membrane raft, membrane microdomain (cellular component); ubiquitin-like protein ligase binding, ubiquitin protein ligase binding, protein serine/threonine kinase activity (molecular function) (Figure 5A and B; Supplementary Table S2). In KEGG enrichment analysis, the differentially expressed autophagy-related genes mainly involved in the process of autophagy and mitophagy (Figure 6; Supplementary Table S3).
 |
Figure 6 Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of 40 differentially expressed autophagy-related genes. |
Validation the Differentially Expressed Autophagy-Related Genes in COPD Patients – Prospective Analysis in New Population
To validate the reliability of the GSE38974 dataset, the expression levels of top five differentially expressed autophagy-related genes were further identified by qRT-PCR in our clinical samples. The clinicopathological variables of cases and controls are summarized in Table 2. Similar to the results of mRNA microarray in lung tissue samples, the expression levels of HIF1A, CDKN1A and BAG3 were significantly higher in COPD blood samples than in normal blood samples (Figure 7A). In addition, the expression levels of ERBB2 and ATG16L1 were significantly decreased (Figure 7B). However, the expression levels of NAF1, CHMP4B, GAPDH, FAM48A and ITPR1 showed no significant difference between the two groups (Figure 7A and B).
 |
Table 2 Clinicopathological Variables of Cases and Controls in This Study |
Discussion
The development of COPD is influenced by smoking, inflammation, airway remodeling and other factors. Increasing evidences suggested that autophagy may be involved in the pathogenesis of COPD. Smoking can lead to oxidative stress of lung cells, which further leads to autophagy activation of lung epithelial cells. Autophagy causes emphysema by inducing autophagic death of lung cells.14 Another evidence suggested that autophagy can promote the release of inflammatory cytokines, which contributes to the inflammatory response of COPD.15 However, extensive validations are needed to improve the understanding of autophagy in pathogenesis of COPD.
A series of recent studies have explored the link between PBMCs and lung tissue RNAs and COPD. Dang et al performed RNA profiling of PBMCs from smokers and COPD patients by microarray. In their study, some differentially expressed miRNAs and mRNAs including miRNA-320b, miR-24-3p, CD177 and IL6 were identified.16 In addition, a recent study found that circRNA0001859 was down-regulated in lung tissue of mice and may be a potential biomarker for the treatment of COPD.17 Moreover, one previous study demonstrated that autophagy‑associated protein levels of PBMCs in COPD patients were increased and were correlated with FEV1% predicted values and circulating levels of cytokines.15 However, there are still few related studies in this field, and further investigations will be required to better understand this field.
To the best of our knowledge, there are several published cancer-related articles exploring autophagy-related genes.18–20 For instance, a recent study reported 30 autophagy-related genes in lung adenocarcinoma, some of which are associated with the prognosis of patients.19 However, bioinformatics analysis of autophagy-related genes has not been explored in COPD. In this study, we were the first identified 40 potential autophagy-related genes in COPD through bioinformatics analysis. Some of these autophagy-related genes of COPD has been previously studied. For example, Ding et al demonstrated that the serum VEGFA is one promising diagnostic biomarker of asthma-COPD overlap syndrome.21 In addition, evidence suggested that SIRT1 and FOXO1 mRNA expression levels in PBMCs correlate to physical activity in COPD patients.22 We intend to explore more potential autophagy-related genes of COPD in the future.
The potential biological functions of these differentially expressed autophagy-related genes were also conducted through GO and KEGG enrichment analysis. GO and KEGG enrichment analysis of differentially expressed autophagy-related genes indicated several enriched terms related to autophagy and mitophagy. Several published articles have confirmed that autophagy can affect the progress of COPD. One in vitro study revealed that miR-21 could increase autophagy and promote the apoptosis of 16HBE cells in COPD.23 Another recent study showed that FUNDC1 silencing could suppress the progress of COPD by inhibiting mitochondrial autophagy through interaction with DRP1.24 Future experiments will be necessary to explore the potential biological functions of these differentially expressed autophagy-related genes.
Based on bioinformatics analysis results, the expression levels of top five differentially expressed autophagy-related genes were further identified by qRT-PCR in our clinical samples. The results of qRT-PCR showed that the expression levels of HIF1A, CDKN1A, BAG3, ERBB2 and ATG16L1 were consistent with the bioinformatics analysis results from mRNA microarray. Some of these verified genes have been reported to be associated with respiratory diseases. Wang et al revealed that the HIF1A gene rs10873142 polymorphism increases the risk of COPD in a Chinese Han population.25 Another study also found that the expression level of CDKN1A is significantly higher in COPD samples when compared with normal samples, in agreement with the results from our clinical samples.26 BAG3 has not been reported in COPD, but BAG3 could promote resistance to apoptosis through Bcl-2 family members in non-small cell lung cancer (NSCLC).27 One evidence demonstrated that the rs4719839 SNP in ATG16L1 can influence the expression of miR-148 and negatively regulate ATG16L1 expression to increase the risk of ventilator-associated pneumonia.28 However, the explicit mechanism of these verified genes in COPD remains largely unclear and needs to be further explored.
Several studies have reported that autophagy is involved in different respiratory diseases. For instance, one research demonstrated that YBX1 (Y-box binding protein 1) could mediate autophagy by targeting p110β and decreasing the sensitivity to cisplatin in NSCLC.29 Moreover, miR-30a targets ATG5 (autophagy-related 5) and attenuates airway fibrosis in asthma by suppressing autophagy.30 Recent study reported a novel role of CX3CR1 (C-X3-C motif chemokine receptor 1) in regulation of macrophage autophagy and promotion of pulmonary fibrosis in hyperoxic lung injured mice.31 In summary, autophagy plays an important role in respiratory diseases.
There are still some limitations in our research. First, the bioinformatics results were obtained from the lung tissues of smokers and COPD patients. However, we performed experimental verification in blood samples of the included individuals. Second, the number of clinical samples included in our study is limited, and we need to confirm our conclusions in a larger COPD cohort. Third, we only verified the expression level of the differentially expressed autophagy-related genes in clinical samples, but did not explore the potential mechanism of these genes in COPD cells and mouse models. Therefore, further research needs to be explored in the future.
Conclusion
In conclusion, we identified 40 potential autophagy-related genes of COPD through bioinformatics analysis. Moreover, the key genes HIF1A, CDKN1A, BAG3, ERBB2 and ATG16L1 may affect the development of COPD by regulating autophagy. These results expanded our understanding of COPD and might be useful in treatment of COPD.
Abbreviations
BP, biological process; BMI, body mass index; CC, cellular component; COPD, chronic obstructive pulmonary disease; GEO, gene expression omnibus dataset; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; MF, molecular function; NSCLC, non-small cell lung cancer; PBMCs, peripheral blood mononuclear cells; PCA, principal component analysis; PPI, protein–protein interactions; qRT-PCR, quantitative real-time polymerase chain reaction; SD, standard deviation.
Ethics Approval and Informed Consent
All procedures performed in studies involving humans were reviewed and permitted by the Tianjin Medical University General Hospital. Written informed consent was obtained from all individual participants included in the study. The study was conducted in accordance with the Declaration of Helsinki.
Consent for Publication
All participating authors give their consent for this work to be published.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 81670084 and No.81970084); and the Tianjin Key Research and Development Program (No. 20YFZCSY00390); and the General Hospital of Tianjin Medical University Youth Incubation Foundation (No. ZYYFY2018030).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Rabe KF, Watz H. Chronic obstructive pulmonary disease. Lancet. 2017;389(10082):1931–1940. doi:10.1016/S0140-6736(17)31222-9
2. Silvestri GA, Young RP. Strange bedfellows: the interaction between COPD and lung cancer in the context of lung cancer screening. Ann Am Thorac Soc. 2020;17(7):810–812. doi:10.1513/AnnalsATS.202005-433ED
3. Yuan C, Chang D, Lu G, Deng X. Genetic polymorphism and chronic obstructive pulmonary disease. Int J Chronic Obstr. 2017;12:1385–1393.
4. Hikichi M, Mizumura K, Maruoka S, Gon Y. Pathogenesis of chronic obstructive pulmonary disease (COPD) induced by cigarette smoke. J Thorac Dis. 2019;11(Suppl S17):S2129–S2140. doi:10.21037/jtd.2019.10.43
5. Csoma B, Bikov A, Nagy L, et al. Dysregulation of the endothelial nitric oxide pathway is associated with airway inflammation in COPD. Resp Res. 2019;20(1):156. doi:10.1186/s12931-019-1133-8
6. Zheng M, Hong W, Gao M, et al. Long noncoding RNA COPDA1 promotes airway smooth muscle cell proliferation in chronic obstructive pulmonary disease. Am J Resp Cell Mol. 2019;61(5):584–596. doi:10.1165/rcmb.2018-0269OC
7. Zong D, Li J, Cai S, et al. Notch1 regulates endothelial apoptosis via the ERK pathway in chronic obstructive pulmonary disease. Am J Physiol Cell Physiol. 2018;315(3):C330–C340. doi:10.1152/ajpcell.00182.2017
8. Gouzi F, Blaquière M, Catteau M, et al. Oxidative stress regulates autophagy in cultured muscle cells of patients with chronic obstructive pulmonary disease. J Cell Physiol. 2018;233(12):9629–9639. doi:10.1002/jcp.26868
9. Galluzzi L, Baehrecke EH, Ballabio A, et al. Molecular definitions of autophagy and related processes. EMBO J. 2017;36(13):1811–1836. doi:10.15252/embj.201796697
10. Huang T, Wan X, Alvarez AA, et al. MIR93 (microRNA −93) regulates tumorigenicity and therapy response of glioblastoma by targeting autophagy. Autophagy. 2019;15(6):1100–1111. doi:10.1080/15548627.2019.1569947
11. Xia F, Deng C, Jiang Y, et al. IL4 (interleukin 4) induces autophagy in B cells leading to exacerbated asthma. Autophagy. 2018;14(3):450–464. doi:10.1080/15548627.2017.1421884
12. Zhang F, Ma H, Wang ZL, Li WH, Liu H, Zhao YX. The PI3K/AKT/mTOR pathway regulates autophagy to induce apoptosis of alveolar epithelial cells in chronic obstructive pulmonary disease caused by PM2.5 particulate matter. J Int Med Res. 2020;48(7):1220727471.
13. Ezzie ME, Crawford M, Cho JH, et al. Gene expression networks in COPD: microRNA and mRNA regulation. Thorax. 2012;67(2):122–131. doi:10.1136/thoraxjnl-2011-200089
14. Lv X, Li K, Hu Z. Chronic obstructive pulmonary disease and autophagy. Adv Exp Med Biol. 2020;1207:559–567.
15. Wu Y, Xu B, He X, et al. Correlation between autophagy levels in peripheral blood mononuclear cells and clinical parameters in patients with chronic obstructive pulmonary disease. Mol Med Rep. 2018;17(6):8003–8009. doi:10.3892/mmr.2018.8831
16. Dang X, Qu X, Wang W, et al. Bioinformatic analysis of microRNA and mRNA Regulation in peripheral blood mononuclear cells of patients with chronic obstructive pulmonary disease. Resp Res. 2017;18(1):4. doi:10.1186/s12931-016-0486-5
17. Chen S, Yao Y, Lu S, et al. CircRNA0001859, a new diagnostic and prognostic biomarkers for COPD and AECOPD. BMC Pulm Med. 2020;20(1):311. doi:10.1186/s12890-020-01333-1
18. Wang SS, Chen G, Li SH, et al. Identification and validation of an individualized autophagy-clinical prognostic index in bladder cancer patients. Oncotargets Ther. 2019;12:3695–3712. doi:10.2147/OTT.S197676
19. Wang X, Yao S, Xiao Z, et al. Development and validation of a survival model for lung adenocarcinoma based on autophagy-associated genes. J Transl Med. 2020;18(1):149. doi:10.1186/s12967-020-02321-z
20. Yang H, Han M, Li H. Construction and validation of an autophagy-related prognostic risk signature for survival predicting in clear cell renal cell carcinoma patients. Front Oncol. 2020;10:707. doi:10.3389/fonc.2020.00707
21. Ding Q, Sun S, Zhang Y, et al. Serum IL-8 and VEGFA are two promising diagnostic biomarkers of asthma-COPD overlap syndrome. Int J Chronic Obstr. 2020;15:357–365.
22. Taka C, Hayashi R, Shimokawa K, et al. SIRT1 and FOXO1 mRNA expression in PBMC correlates to physical activity in COPD patients. Int J Chronic Obstr. 2017;12:3237–3244.
23. Zeng Z, He S, Lu J, et al. MicroRNA-21 aggravates chronic obstructive pulmonary disease by promoting autophagy. Exp Lung Res. 2018;44(2):89–97. doi:10.1080/01902148.2018.1439548
24. Wen W, Yu G, Liu W, et al. Silencing FUNDC1 alleviates chronic obstructive pulmonary disease by inhibiting mitochondrial autophagy and bronchial epithelium cell apoptosis under hypoxic environment. J Cell Biochem. 2019;120(10):17602–17615. doi:10.1002/jcb.29028
25. Wang L, Tang Y, Chen Y. HIF1A gene rs10873142 polymorphism is associated with risk of chronic obstructive pulmonary disease in a Chinese Han population: a case-control study. Biosci Rep. 2018;38(2).
26. Yang D, Yan Y, Hu F, Wang T. CYP1B1, VEGFA, BCL2, and CDKN1A affect the development of chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis. 2020;15:167–175. doi:10.2147/COPD.S220675
27. Zhang Y, Wang JH, Lu Q, Wang YJ. Bag3 promotes resistance to apoptosis through Bcl-2 family members in non-small cell lung cancer. Oncol Rep. 2012;27(1):109–113. doi:10.3892/or.2011.1486
28. Wang SP, Li W, Li C, Duan XY, Duan J. Effect of rs4719839 polymorphism on risk of ventilator-associated pneumonia, expression of microRNA-148 and autophagy-related 16-like 1 (ATG16L1). J Cell Mol Med. 2020.
29. Cui Y, Li F, Xie Q, et al. YBX1 mediates autophagy by targeting p110β and decreasing the sensitivity to cisplatin in NSCLC. Cell Death Dis. 2020;11(6):476. doi:10.1038/s41419-020-2555-4
30. Li BB, Chen YL, Pang F. MicroRNA-30a targets ATG5 and attenuates airway fibrosis in asthma by suppressing autophagy. Inflammation. 2020;43(1):44–53. doi:10.1007/s10753-019-01076-0
31. Chen Y, Zhang H, Li F, Wang X. Inhibition of CX3C receptor 1-mediated autophagy in macrophages alleviates pulmonary fibrosis in hyperoxic lung injury. Life Sci. 2020;259:118286. doi:10.1016/j.lfs.2020.118286
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.