Back to Journals » Cancer Management and Research » Volume 13
HSPA4 Knockdown Retarded Progression and Development of Colorectal Cancer
Authors Zhang M, Dai W, Li Z, Tang L, Chen J, Chen C
Received 16 March 2021
Accepted for publication 14 May 2021
Published 14 June 2021 Volume 2021:13 Pages 4679—4690
DOI https://doi.org/10.2147/CMAR.S310729
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chien-Feng Li
Mingliang Zhang,* Weigang Dai,* Zhanyu Li, Liang Tang, Jianhui Chen, Chuangqi Chen
Department of Colorectal Surgery, Gastrointestinal Surgery Center, The First Affiliated Hospital of Sun Yat-Sen University, Guangzhou, 510080, Guangdong Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chuangqi Chen
Department of Colorectal Surgery, Gastrointestinal Surgey Center, The First Affiliated Hospital of Sun Yat-Sen University, Guangzhou, 510080, Guangdong Province, People’s Republic of China
Email [email protected]
Purpose: Colorectal cancer (CRC) is a common malignancy associated with high morbidity and mortality. Heat shock 70 kDa protein 4 (HSPA4) has been shown to exert regulatory roles during tumor progression in different cancer types. Here, we investigated the expression and cellular functions of HSPA4 in CRC.
Materials and Methods: Expression of HSPA4 in CRC tissues and paracancerous tissues was analyzed by RT-qPCR and immunohistochemistry IHC staining. The functional roles of HSPA4 were explored using shRNA-mediated knockdown in HCT116 and RKO CRC cell lines, both in vitro and in tumor xenograft studies.
Results: HSPA4 expression was significantly increased at the RNA and protein levels in CRC tissues compared with noncancerous tissues. Moreover, HSPA4 expression was positively associated with tumor stage and its high expression of HSPA4 indicated poor patient prognosis. In vitro studies established that HSPA4 knockdown inhibited proliferation and migration, causing arrest in the G2-phase of the cell cycle along with increased levels of apoptosis. This phenotype was recapitulated in vivo where HSPA4 knockdown suppressed xenograft growth. Mechanistic investigations showed silencing of HSPA4 reduced activation of the PI3K, Akt signaling axis while also downregulating the cell cycle progression markers, CCND1 and CDK6. Similarly, there was altered expression of apoptosis-related proteins consistent with the increase in apoptosis.
Conclusion: Our findings demonstrate clinical significance for HSPA4 in CRC, further showing that HSPA4 contributes to CRC tumorigenesis through effects on proliferation, migration and survival. Thus, HSPA4 represents a novel prognostic indicator as well as a promising therapeutic target in CRC.
Keywords: CRC, HSPA4, proliferation, apoptosis, migration
Introduction
Colorectal cancer (CRC) occurs at high incidence and represents the fourth leading cause of cancer deaths worldwide.1 Projections to the year 2035 indicate CRC prevalence will increase by 60%, accounting for 2.2 million new annual cases and over 1.1 million annual deaths.2 Current CRC treatments typically involve surgery and radiotherapy, as well as the use of chemotherapeutic drugs such as FOLFOX and FOLFIRI regimens.3 Additionally to improve outcomes, chemotherapy is now often combined with monoclonal antibodies or other targeted therapies, eg against vascular endothelial growth factor (VEGF) or epidermal growth receptor (EGFR).4 Despite such advancements, recurrence rates for CRC are high, resulting in poor survival outcomes.5 Consequently, there is an urgent need to thoroughly understand the molecular mechanisms of CRC and identify key molecular targets, thus laying the foundation for the development of new and more effective treatments.
Heat shock proteins (HSPs) are ubiquitous cellular molecules that act as a molecular chaperones under different stress conditions, including carcinogenesis.6 Previous studies have found a highly abundant set of HSPs on the surface of tumor cells, including HSPA4, HSP60, HSP70, HSP90, and HSP27.6 HSPA4 (heat shock protein 4) is a 70kDa member of the HSP110 family7 and there is emerging evidence to suggest that HSPA4 participates in the progression and development of different cancers. For example, Jo et al reported frameshift mutations in HSPA4 in both gastric cancer and CRC8 and moreover, Wang et al showed that HSPA4 may be important for nasopharyngeal carcinoma (NPC) metastasis.9 Additionally, Ma et al found significant correlations between HSPA4 expression and overall survival in hepatocellular carcinoma (HCC) patients.10 Moreover, Gu et al proposed that tumor-educated B cells selectively promote breast cancer lymph node metastasis by HSPA4-targeting IgG.11 Interestingly, Han et al inferred that inhibition of LPS-induced cardiomyocyte apoptosis and mitochondrial damage by suppressing post-transcriptional regulation of HSPA4 by miR-1-5p.12 Although HSPA4 has been implicated in various biological processes related to tumorigenesis, its cellular functions and molecular mechanisms in CRC are still largely unknown.
The aim of this study was to elucidate the relationship between the expression of HSPA4 and CRC tumorigenicity. To this end, we identified that HSPA4 was commonly upregulated in CRC and its expression was associated with poor prognosis. Functional interrogation using in vitro assays indicated that HSPA4 knockdown inhibited cell proliferation and suppressed migration also causing cell cycle arrest and promoting cell apoptosis. Consistently, assessment of the effects of HSPA4 knockdown in vivo also showed inhibition of tumor growth. Our findings suggest that HSPA4 functionally contributes to the development and progression of CRC and moreover propose that HSPA4 may be a therapeutic target.
Materials and Methods
CRC Tissue Samples and Cell Lines
A total of 87 tumor tissues and 69 noncancerous tissues were collected from CRC patients at the First Affiliated Hospital of Sun Yat-sen University. None of the patients received radiotherapy or chemotherapy prior to biopsy. This study was approved by the Research Ethics Committee of the First Affiliated Hospital of Sun Yat-sen University with written informed consent was obtained from each participant.
The human HCT116 and RKO CRC cell lines were purchased from Cell Bank of the Chinese Academy of Sciences (Shanghai, China) in August 2019 and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C in a 5% CO2 atmosphere. All cells were authenticated by STR and confirmed negative for mycoplasma contamination prior to the experiments (Figure S1A-B).
Immunohistochemical (IHC) Staining
Tissue sections were deparaffinized and subjected to antigen retrieval with citric acid before incubation with primary antibodies (anti-HSPA4, 1:200, # ab185962, Abcam USA) and then IgG HRP-conjugated secondary antibody (1:400, # ab6721, Abcam). Incubation steps were performed at room temperature for 3 h and afterwards antibody complexes were detected with DAB and the tissue sections counterstained with hematoxylin. Specimens were independently evaluated by two pathologists with IHC scores expressed as staining percentage defined as: 1 (1–24%), 2 (25–49%), 3 (50–74%), 4 (75–100%) and staining intensity defined as 0: no color, 1: brown, 2: light yellow, 3: dark brown. Accordingly, high or low expression groups for HSPA4 were defined by median IHC scores.
RNA Interference
Short hairpin RNAs (shRNA) against human HSPA4 or a related control sequence was designed and inserted into the BR-V-108 lentiviral plasmid vector by Shanghai biosciences Co., Ltd. (Shanghai, China). The target sequences used were shHSPA4-1: 5ʹ-AAAGTCAAAGTTCGAGTAAAT-3ʹ; shHSPA4-2: 5ʹ-GGTGACATATATGGAGGAAGA-3ʹ; shHSPA4-3: 5ʹ- AAGCAATGGAGTGGATGAATA-3ʹ. Plasmids were packaged using 293T cells using plasmids prepared using the EndoFree Maxi Plasmid Kit (Tiangen). HCT116 or RKO cells were seeded at a density of 2×105 cells/mL in a 6-well plate and infected after 24 h with 100 μL lentiviral supernatant (1×107 TU/mL) along with ENI.S and polybrene (10 µg/mL, Sigma-Aldrich). After 72 h, cell infection was verified by fluorescence microscopy and knockdown efficiency determined via qRT-PCR and Western blotting.
RNA Isolation and Quantitative Real Time-Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from the tumor and noncancerous tissues using the Trizol reagent (Invitrogen, CA, USA) according to the procedure manufacturers protocol. Total RNA was similarly extracted from CRC cell lines. RNA (2.0 μg) was transcribed into cDNA using the M-MLV RT kit (Promega) and triplicate qRT-PCR reactions performed on the Roche Light Cycler® 96 real-time PCR platform. The expression of HSPA4 was quantified using the 2−ΔΔCT method using GAPDH as the internal control. The primer sequences used were: HSPA4: 5ʹ- GCAGACACCAGCAGAAAATAAGG-3ʹ, 5ʹ- TCGATTGGCAGGTCCACAGT-3ʹ; GAPDH: 5ʹ- TGACTTCAACAGCGACACCCA-3ʹ, 5ʹ-CACCCTGTTGCTGTAGCCAAA-3ʹ.
Western Blotting
Cell lysates were prepared from shCtrl or shHSPA4 infected HCT116 and RKO cell lines were lysed in ice-cold RIPA buffer (Millipore) and. The protein concentrations determined using the BCA Protein Assay Kit (#23225, HyClone-Pierce). Equal protein amounts (20 μg) were separated by 10% SDS-PAGE, transferred onto PVDF membranes, and blotted with primary antibodies against HSPA4 (1:2000, Abcam, #ab185962), Akt (1:1000, CST, #4685), p-Akt (1:1000, Bioss, #BS-5193R), CCND1 (1:2000, CST, #2978), CDK6 (1:1000, Abcam, #ab151247), PIK3CA (1:1000, Abcam, #ab40776) and GAPDH (1:3000, Bioworld, #AP0063). The blots were visualized with the Amersham ECL plusTM Western blotting system and the density of protein bands was analyzed using ImageJ software (National Institute of Health).
Celigo Cell Counting Assay
The shCtrl or shHSPA4 infected HCT116 and RKO cell lines were seeded into 96-well plates (500 cells/well). After culturing to allow cell clones to form, the wells were fixed with 4% formaldehyde and then with Giemsa. Visible colonies were then photographed and counted for 5 consecutive days.
Flow Cytometry
HCT116 and RKO cells infected with shCtrl or shHSPA4 lentivirus were cultured until cell density reached 85% before harvesting and analysis. Thereafter, apoptosis analyses and cell cycle distribution assays were performed using a Guava easyCyte HT FACS Calibur flow cytometer (Millipore). Briefly, 10 μL Annexin V-APC (eBioscience) was added to cells suspended in 800 μL 1× binding buffer and incubated at room temperature without light for 10 min. The cell apoptotic ratio was calculated based on the following formula: (number of positive cells/numbers of all counted cells) × 100%. For cell cycle analyses, the cells were stained using PI staining solution (BD Biosciences) and ratio of cells in the G1, S and G2 cell cycle phases determined by flow cytometry.
Wound Healing Assay
Cell migration was measured by scratch wound healing assays. HCT116 and RKO cells infected with shCtrl or shHSPA4 lentivirus were plated into 96-well plates (5×104 cells/well) and cultured until cell confluence reached 90%. Subsequently, cell scratches were made across the cell layer by a 96-wounding replicator (VP scientific) and the cell layers gently washed. Photographs were taken using epifluorescence microscopy at 24 h and 48 h post scratching and the migration rates calculated.
Transwell Assay
HCT116 and RKO cells infected with shCtrl or shHSPA4 lentivirus were seeded at 8×104 cells in the upper chamber of 24-well transwell cell culture plates. The lower chamber was filled with 600 μL medium supplemented with 30% FBS. The non-migratory cells on the upper chamber were removed with a cotton swab, while the cells adhering to the membrane were fixed in 4% paraformaldehyde (precooled with ice) for 30 min and stained with 0.1% crystal violet for 20 min at room temperature. Following washing with PBS, 5 fields of view per well were selected randomly under a 200 ×magnification microscope, and images were captured for enumeration of the cells.
Xenograft Model
BALB/c nude mice (4 weeks old) obtained from Shanghai Lingchang Laboratory Animal Technology Co., Ltd. Ten mice were randomly selected and divided into the shCtrl (n=5) and shHSPA4 (n=5) groups. A total of 4×106 RKO cells infected with shCtrl or shHSPA4 lentivirus were subcutaneously injected into the right forearm armpit of each mouse. After 6 days of injection, the tumor size and mice weight were monitored 2 times per week, and tumor volume =π/6× L × W2, where L represents the long diameter and W represents the short diameter. In vivo bioluminescence images were also collected using the Berthold Technologies living imaging system on mice anesthetized by intraperitoneal injection of 0.7% Pentobarbital Sodium (10 uL/g). Subsequently, mice were sacrificed and the tumor tissues were harvested. All animal experiments were approved by the Ethics committee of the First Affiliated Hospital of Sun Yat-sen University and conducted in accordance with Guide for Care and Use of Laboratory animals (NIH publication number 85–23, revised at 1996).
Ki67 Staining
Xenografted tumor tissues were fixed in 10% formalin and embedded in paraffin. After preparing 5 μm sections, the tissues were dewaxed in xylene and ethanol before rehydration. Tissue sections were blocked with 3% PBS-H2O2 and were incubated with anti-Ki67 and HRP goat anti-rabbit IgG. Alternatively, slides were stained by Hematoxylin (Baso, # BA4041) and Eosin (Baso, # BA4022). Stained slides were examined at 200× and 400× by light microscopy.
Human Apoptosis Antibody Array
The Human Apoptosis Antibody Array (Abcam, # ab134001) was used to quantitate signal pathway protein detection following the manufacturer’s instructions. Briefly, cell lysates were prepared as for Western blotting and equal protein amounts incubated with pre-blocked array antibody membranes overnight at 4°C. After washing, 1:100 Detection Antibody Cocktail was incubated for 1 h, followed by incubation with HRP linked streptavidin conjugate for 1 h. All spots were visualized by enhanced ECL and the signal densities were analyzed with ImageJ software (National Institute of Health).
Statistical Analysis
All experiments were performed in triplicate and data shown as mean ± SDs. Statistical analyses and graphs were performed by GraphPad Prism 8.01 (GraphPad Software) with P value < 0.05 considered to be statistically significant. Significance differences between groups were determined using the two-tailed Student’s t test or One-way ANOVA. The relationships between HSPA4 expression and tumor characteristics in patients with CRC was analyzed by multivariate cox regression analysis.
Results
Upregulation of HSPA4 is Associated with Poor Prognosis in CRC Patients
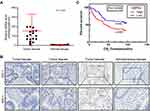
To evaluate the role of HSPA4 in CRC, we first examined whether the expression level of HSPA4 was altered in CRC compared to normal tissues. Examination of mRNA levels using qRT-PCR indicated that HSPA4 was higher expressed in CRC tissues compared to noncancerous tissues (P < 0.01) (Figure 1A). Moreover, these changes were confirmed at the protein level using IHC (Figure 1B) (Table 1). Furthermore, the relationship between HSPA4 expression and tumor characteristics indicated a significant positive correlation between HSPA4 expression and tumor stage (P = 0.019) (Table 2). Subsequent multivariate cox regression analysis indicated that the expression levels of HSPA4 were positively correlated with tumor differentiation (P = 0.014) and stage (P = 0.041) (Table 3). Most instructively, Kaplan–Meier survival analysis revealed that HSPA4 expression levels were significantly associated with overall survival in CRC patients (Figure 1C). Collectively these results highlight that high expression of HSPA4 has the clinical significance in CRC, being associated with tumor malignancy and of indicating poor prognosis in patients.
 |
Table 1 Expression Patterns in Colorectal Cancer Tissues and Para-Carcinoma Tissues Revealed in Immunohistochemistry Analysis |
 |
Table 2 Relationship Between HSPA4 Expression and Tumor Characteristics in Patients with Colorectal Cancer |
 |
Table 3 The Correlation Between HSPA4 Expression and Multivariate Cox Regression |
ShRNA-Mediated Knockdown of HSPA4 in CRC Cell Lines
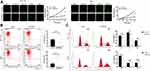
Analysis of mRNA expression in CRC cell lines using qRT-PCR indicated that HSPA4 levels in HCT116 and RKO cells were more abundant than other cell lines tested (Figure S2A). To assess the function of HSPA4 in CRC, we then constructed loss-of-function cell models in HCT116 and RKO cells using shRNA-mediated gene silencing. Evaluation of three independent targeting sequences using qRT-PCR showed that shHSPA4-3 produced the highest knockdown efficiency (Figure S2B), and this vector was chosen for subsequent experiments. HCT116 and RKO cells were then infected with shHSPA4 or a control sequence (shCtrl) with epifluorescence imaging showing that the infection efficiency reached over 80% in 72 h (Figure S2C). Moreover, qRT-PCR analysis revealed that HSPA4 knockdown efficiencies were 96.1% (P < 0.01) and 94.94% (P < 0.01) respectively, in HCT116 and RKO cells were compared with the shCtrl control (Figure S2D). These findings were then verified at the protein level with Western blotting results showing that the HSPA4 protein expression after HSPA4 knockdown was significantly reduced compared to the shCtrl group in both HCT116 and RKO cells (Figure S2E). Together these findings indicate robust knockdown of HSPA4 in the HCT116 and RKO cell models.
Knockdown of HSPA4 Inhibits Proliferation of HCT116 and RKO Cells in vitro
We next used Celigo cell counting assays using the HCT116 and RKO cell models to determine if HSPA4 expression affected cell proliferation. Indeed, cell count observations indicated that the cell proliferation ratio of the shHSPA4 groups was reduced compared to the shCtrl groups for both HCT116 and RKO cells, with growth rates only about one-third after 5 days of culture (P < 0.001) (Figure 2A). These experimental results indicate that decreasing HSPA4 expression in CRC cells lines can significantly inhibit their proliferation.
Knockdown of HSPA4 Promotes Apoptosis and Cell Cycle Arrest in HCT116 and RKO Cells in vitro
To further investigate the growth inhibitory phenotype observed after HSPA4 knockdown, we employed flow cytometry to measure apoptotic rates as well as cell cycle parameters in the CRC cell models. Notably, compared with shCtrl group, the apoptotic rates increased after HSPA4 knockdown, by factors of ~2 and ~3.3 fold in HCT116 and RKO cells, respectively (P < 0.001) (Figure 2B). Furthermore, the numbers of HCT116 and RKO cells in the G2 phase were increased in the shHSPA4 compared to the shCtrl groups (P < 0.01) (Figure 2C). In contrast, the proportion of S phase cells were decreased for HCT116 cells while this increased in RKO cells. The cause of this difference is unclear but nonetheless, HSPA4 knockdown is associated with cell cycle arrest in the G2 phase. Therefore, HSPA4 knockdown increased apoptosis and promotes cell cycle arrest in CRC cells in vitro.
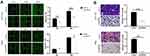
Knockdown of HSPA4 Inhibits Migration of HCT116 and RKO Cells in vitro
Next, wound healing and Transwell assays were used to determine the effects of HSPA4 knockdown on CRC cell motility. Wound healing assays showed the migration rate of HCT116 shHSPA4 cells was reduced by 39% compared to shCtrl controls over 48 h (P < 0.001) (Figure 3A). Interestingly, no significant alterations in RKO cell migration were caused by HSPA4 knockdown after 30 h. However, the effects of HSPA4 downregulation on CRC cell migration were more consistent in Transwell assays. Here, the migration rates of HCT116 and RKO cells in the shHSPA4 group were reduced by about 60% and 90%, respectively, compared with the shCtrl group (P < 0.01) (Figure 3B). Collectively these findings indicate that HSPA4 knockdown inhibits the migration of CRC cells.
Knockdown of HSPA4 in CRC Cells Impairs Tumorigenesis in vivo
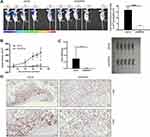
Based on our in vitro findings that HSPA4 knockdown inhibited cell proliferation and migration while promoting apoptosis, we anticipated that HSPA4 knockdown would also inhibit CRC tumorigenesis in vivo. To test this notion, RKO cells with or without HSPA4 knockdown were subcutaneously injected into nude mice to establish a mouse xenotransplantation model. As anticipated, bioluminescence imaging showed that tumor growth in shHSPA4 bearing tumors was significantly slower than shCtrl controls (P < 0.001) (Figure 4A). Comparisons of tumor volume measurements showed the shHSPA4 group was smaller than shCtrl group 18 days after injection (302.45 ± 266.26 mm3 versus, P < 0.05) (Figure 4B). The mean tumor weight at the conclusion of the experiment in the shHSPA4 group was 0.29g ± 0.269 g lighter than the shCtrl group (P < 0.05) (Figure 4C and D). Additionally, examination of the proliferation index of tumor tissues using Ki67 staining showed that proliferation in shHSPA4 tumors was significantly lower than that in the shCtrl tumors (Figure 4D). These findings were therefore consistent with the in vitro experiments, suggesting that HSPA4 knockdown attenuates CRC tumorigenicity.
Exploration of Signaling Mechanisms Downstream of HSPA4 in CRC Cell
We next used the Human Apoptosis Antibody Array to investigate the signaling changes associated with HSPA4 knockdown in CRC. Using the array, we investigated the differential expression of 43 proteins key apoptotic between the shHSPA4 and shCtrl RKO cells Notably, after HSPA4 silencing the expression of Caspase 3, HSP60, IGFBP-6 and SMAC were significantly upregulated in RKO cells (Figure 5A). Conversely, the expression levels of Bcl-2, Bcl-w, IGF-I, IGF-II and IGF-1sR were downregulated by HSPA4 knockdown (P < 0.05). Additional analysis by Western blotting to investigate other key signaling proteins showed that the expression of p-Akt, CCND1, CDK6, PIK3CA was downregulated in shHSPA4 cells compared to the shCtrl cells, although Akt expression was not significantly changed (Figure 5B). These data suggested that knockdown of HSPA4 reduced the activation of PI3K, Akt signaling, while the downregulation of CCND1 and CDK6 suggests inhibition of cell cycle progression. These changes are consistent with the HSPA4 knockdown phenotype showing decreased cell proliferation.
Discussion
HSPA4 has been reported to participate in diverse biological processes, many which involve tumor progression. Here, we investigated the role of HSPA4 in CRC, finding that HSPA4 is overexpressed in CRC where its high expression levels indicate poor prognosis. In addition, our investigation in two in CRC cell lines (HCT116 and RKO) showed that knockdown of HSPA4 inhibits different malignant characteristics, including proliferation and migration while enhancing apoptosis. Furthermore, the decreased expression of HSPA4 in RKO cells leads to the alterations in apoptosis-related proteins.
Apoptosis is recognized as the most important form of cell death,13 which involves a series of cellular signals.14 Our study found that HSPA4 knockdown upregulated proapoptotic molecules such Caspase 3, HSP60, IGFBP-6 and SMAC, and moreover, decreased the levels of apoptosis inhibitors such as Bcl-2, Bcl-w, IGF-I, IGF-II and IGF-1sR. Caspase 3 is one of most closely related molecules in the process of apoptosis, and is responsible for changes in cell morphology and the fragmentation of DNA.15,16 Moreover, upon death stimuli, HSP60 is released from mitochondria to the cytosol to exert a pro-death function, either through stabilizing Bax, enhancing Caspase 3 activation, or increasing protein ubiquitination.17,18 Qiu et al revealed that IGFBP-6 inhibits the invasion and migration of CRC cells possibly via cell cycle arrest and promoting apoptosis.19 Interesting, multiple preclinical studies have documented the ability of SMAC to either directly induce cell death of cancer cells or trigger cell death.20 Conversely, Bcl-2 inhibits cell apoptosis and is closely associated with the onset and drug resistance of a variety of malignant tumors.21 Bcl-W belongs to the Bcl-2 family and is considered as an anti-apoptotic protein in different types of normal and diseased cells.22 IGFs include IGF-I, IGF-II, relevant receptors IGF-IR and IGF-IIR, and IGF-binding proteins (IGFBPs), all of which exert crucial roles in anti-apoptosis and facilitating cell proliferation.23,24 Han et al, illustrated that the progression of CRC may be associated with elevated expression levels of IGFs-related proteins.25 On this basis, the changes in key apoptotic proteins after HSPA4 knockdown are consistent with the apoptotic phenotype observed. Nonetheless, more work is required to better resolve how the molecular signaling pathways of apoptosis are controlled by HSPA4 in CRC cells.
Additionally, HSPA4 knockdown in RKO cells triggered decreases in phosphorylated Akt and downregulation of PIK3CA, CCND1 and CDK6. Many cancers often exhibit activation of the PI3K/Akt/mTOR signaling pathway through alterations in PI3K or Akt,26 and both such molecules are known to play key roles in regulating CRC cell growth.27 Additionally, Bali et al reported that CCND1 was essential for the pathogenesis and metastasis of CRC and can be used as a prognostic index.28 Besides, Zhang et al highlighted that targeted therapies combined with CDK4/6 inhibitors offer promising treatment options for CRC.29 Therefore, targeting HSPA4 could inhibit the progression of CRC cells through downstream effects on PI3K/Akt, CCND1 and CDK6.
Conclusion
Our study determined that HSPA4 is highly expressed in CRC, and its expression has potential clinical value in predicting poor prognosis of CRC patients. In addition, knockdown of HSPA4 inhibits the malignant biological properties of CRC by reducing proliferation, enhancing apoptosis and weakening migration. In summary, HSPA4 may act as a potential prognostic marker and promising therapeutic target for this lethal disease.
Acknowledgment
Mingliang Zhang and Weigang Dai are the first authors.
Funding
Guangdong Science and Technology Plan Project, No. 2013B021800131, Guangzhou health and medical collaborative innovation major projects, No. 201604020003. Science and Technology Program of Guangzhou, No. 202102080387.
Disclosure
The authors declare no conflict of interest.
References
1. Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467–1480. doi:10.1016/S0140-6736(19)32319-0
2. Araghi M, Soerjomataram I, Jenkins M, et al. Global trends in colorectal cancer mortality: projections to the year 2035. Int J Cancer. 2019;144:2992–3000. doi:10.1002/ijc.32055
3. Sandhu J, Lavingia V, Fakih M. Systemic treatment for metastatic colorectal cancer in the era of precision medicine. J Surg Oncol. 2019;119:564–582. doi:10.1002/jso.25421
4. Van Cutsem E, Cervantes A, Nordlinger B, Arnold D, Group EGW. Metastatic colorectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(Suppl 3):iii1–9. doi:10.1093/annonc/mdu260
5. Marmol I, Sanchez-de-diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal carcinoma: a general overview and future perspectives in colorectal cancer. Int J Mol Sci. 2017;18:197. doi:10.3390/ijms18010197
6. Yang Z, Zhuang L, Szatmary P, et al. Upregulation of heat shock proteins (HSPA12A, HSP90B1, HSPA4, HSPA5 and HSPA6) in tumour tissues is associated with poor outcomes from HBV-related early-stage hepatocellular carcinoma. Int J Med Sci. 2015;12:256–263. doi:10.7150/ijms.10735
7. Easton DP, Kaneko Y, Subjeck JR. The hsp110 and Grp1 70 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaperones. 2000;5:276–290. doi:10.1379/1466-1268(2000)005<0276:thagsp>2.0.co;2
8. Jo YS, Choi MR, Song SY, et al. Frameshift Mutations of HSPA4 and MED13 in Gastric and Colorectal Cancers. Pathol Oncol Res. 2016;22:769–772. doi:10.1007/s12253-016-0070-9
9. Wang Y, Zhao Q, Lan N, Wang S. Identification of methylated genes and miRNA signatures in nasopharyngeal carcinoma by bioinformatics analysis. Mol Med Rep. 2018;17:4909–4916. doi:10.3892/mmr.2018.8487
10. Ma C, Xu T, Sun X, et al. Network pharmacology and bioinformatics approach reveals the therapeutic mechanism of action of baicalein in hepatocellular carcinoma. Evid Based Complement Alternat Med. 2019;2019:7518374. doi:10.1155/2019/7518374
11. Gu Y. Tumor-educated B cells selectively promote breast cancer lymph node metastasis by HSPA4-targeting IgG. Nat Med. 2019;25:312–322. doi:10.1038/s41591-018-0309-y
12. Han Y, Cai Y, Lai X, et al. lncRNA RMRP prevents mitochondrial dysfunction and cardiomyocyte apoptosis via the miR-1-5p/hsp70 Axis in LPS-Induced Sepsis Mice. Inflammation. 2020;43:605–618. doi:10.1007/s10753-019-01141-8
13. Wang RA, Li Q-L, Li Z-S, et al. Apoptosis drives cancer cells proliferate and metastasize. J Cell Mol Med. 2013;17:205–211. doi:10.1111/j.1582-4934.2012.01663.x
14. Soleimani A, Bahreyni A, Roshan MK, et al. Therapeutic potency of pharmacological adenosine receptors agonist/antagonist on cancer cell apoptosis in tumor microenvironment, current status, and perspectives. J Cell Physiol. 2019;234(3):2329–2336. doi:10.1002/jcp.27249
15. McIlwain DR, Berger T, Mak TW. Caspase functions in cell death and disease. Cold Spring Harb Perspect Biol. 2013;5:a008656. doi:10.1101/cshperspect.a008656
16. O’Donovan N. Caspase 3 in breast cancer. Clin Cancer Res. 2003;9:738–742.
17. Wyzewski Z, Gregorczyk KP, Szczepanowska J, Szulc-Dabrowska L. Functional role of Hsp60 as a positive regulator of human viral infection progression. Acta Virol. 2018;62:33–40. doi:10.4149/av_2018_104
18. Huang YH, Yeh CT. Functional Compartmentalization of HSP60-survivin interaction between mitochondria and cytosol in cancer cells. Cells. 2019;9:23. doi:10.3390/cells9010023
19. Qiu F, Gao W, Wang B. Correlation of IGFBP-6 expression with apoptosis and migration of colorectal carcinoma cells. Cancer Biomark. 2018;21:893–898. doi:10.3233/CBM-170947
20. Fulda S. Smac mimetics to therapeutically target IAP proteins in cancer. Int Rev Cell Mol Biol. 2017;330:157–169. doi:10.1016/bs.ircmb.2016.09.004
21. Knight T, Luedtke D, Edwards H, Taub JW, Ge Y. A delicate balance - The BCL-2 family and its role in apoptosis, oncogenesis, and cancer therapeutics. Biochem Pharmacol. 2019;162:250–261. doi:10.1016/j.bcp.2019.01.015
22. Hartman ML, Czyz M. BCL-w: apoptotic and non-apoptotic role in health and disease. Cell Death Dis. 2020;11:260. doi:10.1038/s41419-020-2417-0
23. Sharon C, Baranwal S, Patel NJ, et al. Inhibition of insulin-like growth factor receptor/AKT/mammalian target of rapamycin axis targets colorectal cancer stem cells by attenuating mevalonate-isoprenoid pathway in vitro and in vivo. Oncotarget. 2015;6:15332–15347. doi:10.18632/oncotarget.3684
24. Peters G, Gongoll S, Langner C, et al. IGF-1R, IGF-1 and IGF-2 expression as potential prognostic and predictive markers in colorectal-cancer. Virchows Arch. 2003;443:139–145. doi:10.1007/s00428-003-0856-5
25. Han X, Hou S, Yang A. Correlation Between IGFs-Related proteins expression and incidence of colorectal cancer in diabetic patients and related mechanisms. Med Sci Monit. 2016;22:848–854. doi:10.12659/msm.895837
26. Phyu SM, Smith TAD. Combination treatment of cancer cells with pan-Akt and pan-mTOR inhibitors: effects on cell cycle distribution, p-Akt expression level and radiolabelled-choline incorporation. Invest New Drugs. 2019;37:424–430. doi:10.1007/s10637-018-0642-5
27. Slattery ML, Lundgreen A. The influence of the CHIEF pathway on colorectal cancer-specific mortality. PLoS One. 2014;9:e116169. doi:10.1371/journal.pone.0116169
28. Chen WC. Survey of molecular profiling during human colon cancer development and progression by immunohistochemical staining on tissue microarray. World J Gastroenterol. 2007;13:699–708. doi:10.3748/wjg.v13.i5.699
29. Zhang J, Zhou L, Zhao S, Dicker DT, El-Deiry WS. The CDK4/6 inhibitor palbociclib synergizes with irinotecan to promote colorectal cancer cell death under hypoxia. Cell Cycle. 2017;16:1193–1200. doi:10.1080/15384101.2017.1320005
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.