Back to Journals » OncoTargets and Therapy » Volume 13
Hsa_circRNA_100146 Acts as a Sponge of miR-149-5p in Promoting Bladder Cancer Progression via Regulating RNF2
Authors Wang H, Niu X, Mao F, Liu X, Zhong B, Jiang H, Fu G
Received 23 July 2020
Accepted for publication 7 October 2020
Published 29 October 2020 Volume 2020:13 Pages 11007—11017
DOI https://doi.org/10.2147/OTT.S273622
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjay Singh
Hengbing Wang,* Xiaobing Niu,* Fei Mao, Xuzhong Liu, Bing Zhong, Hesong Jiang, Guangbo Fu
Department of Urology, The Affiliated Huaian No. 1 People’s Hospital of Nanjing Medical University, Huai’an 223300, Jiangsu Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Guangbo Fu
Department of Urology, The Affiliated Huaian No. 1 People’s Hospital of Nanjing Medical University, No. 1 West Huanghe Road, Huai’an 223300, Jiangsu Province, People’s Republic of China
Tel +86-517-80872607
Email [email protected]
Background: Mounting evidence has demonstrated that circular RNAs (circRNAs) play indispensable roles in the progression of bladder cancer. Public database mining showed that hsa_circRNA_100146 (circRNA_100146) was highly expressed in bladder cancer. This study aimed to characterize the biological role of circRNA_100146 and clarify the underlying mechanism in bladder cancer.
Methods: We evaluated the relationship between circRNA_100146 expression and clinicopathological features. Furthermore, gain- and loss-of-function studies were conducted in bladder cancer cells via transfection with gene-carrying plasmids (over-expression) or specific short hairpin RNAs (knockdown). Moreover, computational algorithms and dual-luciferase reporter assays were performed to explore the possible mechanisms of action. Additionally, in vivo xenograft experiments were performed to further analyze the effect of circRNA_100146 on tumor growth.
Results: Our data showed that circRNA_100146 expression was increased in bladder cancer tissues and cell lines, and that high expression of circRNA_100146 was correlated with poor patient prognosis. Upregulation of circRNA_100146 promoted cell proliferation, migration, and invasion, and inhibited cell apoptosis, whereas knockdown of circRNA_100146 displayed opposite effects on bladder cancer cells. Notably, circRNA_100146 could combine with miR-149-5p and promote ring finger protein 2 (RNF2) expression, thereby facilitating the progression of bladder cancer. Furthermore, overexpression of RNF2 reversed the effects of circRNA_100146 knockdown on the biological behaviors of bladder cancer cells. The in vivo experiments revealed that downregulation of circRNA_100146 dramatically delayed tumor growth.
Conclusion: Our findings indicate that circRNA_100146 functions as a sponge of miR-149-5p in promoting bladder cancer progression by regulating RNF2 expression and that circRNA_100146 may serve as a novel biomarker in human bladder cancer.
Keywords: bladder cancer, circRNA_100146, proliferation, invasion, RNF2
Introduction
Bladder cancer is the fifth most common cancer worldwide, and it usually occurs in bladder epithelial cells.1 It is estimated that there are about 150,000 new cases of bladder cancer and more than 50,000 bladder cancer-related deaths every year.2 In spite of great advances in treatment, the postoperative recurrence rate is still very high. It is reported that approximately 80% of bladder cancer patients suffer a relapse within 5 years.3 The survival rate of patients with bladder cancer is usually associated with multiple influencing factors, including tumor stage, pathological type, and diagnosis time. Follow-up investigations showed that the 5-year survival rate of patients at stage IV was only 15%.4 It is well known that bladder cancer is a pathological process that is influenced by abnormal expression of numerous genes. Thus, it is urgent to identify molecular markers that are linked to the recurrence and metastasis of bladder cancer.
Circular RNAs (circRNAs) are a novel type of non-coding RNA molecule that plays an indispensable role in diverse types of human diseases. Studies have shown that circRNAs could exert carcinogenic or anti-tumor effects in bladder cancer.5,6 circRNAs are mainly recognized as miRNA molecular sponges that regulate downstream gene expression and signaling pathway transduction, thus affecting the malignant phenotypes of bladder cancer cells.7,8 Several circRNAs have been reported to participate in the occurrence and development of bladder cancer, such as cTFRC,9 circFUT8,10 circMTO1,11 and hsa_circ_0001361.12 Previous studies have shown that has_circRNA_100146 (circRNA_100146), also known as hsa_circ_0011385,12 is highly expressed in non-small cell lung cancer13 and thyroid cancer14 and that circRNA_100146 is an oncogene that promotes the progression of non-small cell lung cancer and thyroid cancer. Based on the GSE92675 dataset analysis, it was found that circRNA_100146 was over-expressed in clinical samples of bladder cancer.15 However, the biological function and molecular mechanism of circRNA_100146 in bladder cancer remain unknown.
In the present study, we investigated the effects of circRNA_100146 on bladder cancer cell proliferation, apoptosis, migration, and invasion. Herein, our data implied that circRNA_100146 may function as an oncogenic circRNA in bladder cancer. Moreover, mechanistic studies revealed that circRNA_100146 and ring finger protein 2 (RNF2) could competitively bind to miR-149-5p to regulate malignant behaviors in bladder cancer cells. Additionally, knockdown of circRNA_100146 was shown to delay the growth of bladder cancer cells in vivo. Taken together, our study provides some evidence for circRNA_100146 as a new biomarker for bladder cancer diagnosis and prognosis.
Materials and Methods
Patient and Tissue Samples
Bladder cancer tissues (n = 68) and matched adjacent non-tumor tissues were obtained from patients who underwent surgical treatment between October 2014 and May 2017 at the Affiliated Huai’an No.1 People’s Hospital of Nanjing Medical University (Huai’an, China). The clinicopathological characteristics of bladder cancer patients are shown in Table 1. Written informed consent was obtained from all patients enrolled in the present study. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Ethics Committee of the Affiliated Huai’an No.1 People’s Hospital of Nanjing Medical University.
 |
Table 1 Correlation Between circRNA_100146 Expression and Clinicopathological Characteristics of Bladder Cancer Patients |
Total RNA Isolation and RNase R Treatment
RNA was extracted from J82 and T24 cells using TRIzol reagent, followed by treatment with DNase I (Ambion) (37 °C, 30 min, twice). Approximately 3 μg of total RNA was extracted from each sample, and ribosomal RNA was removed using the Ribosomal Eukaryotic Kit (Qiagen). The purified RNA was treated with RNase R (Epicenter, 40 U, 37 °C, 3 h) as described previously,16 and then purified using TRIzol.
Quantitative Real-Time PCR (qRT-PCR)
TRIzol reagent (Invitrogen, CA, USA) was used to extract total RNA from cells. RNA was reverse-transcribed using PrimeScript RT Master Mix (Takara, Japan). GAPDH was used as an endogenous control for circRNA_100146 and RNF2, and U6 was used as an endogenous control for miR-149-5p. qRT-PCR was performed under the following conditions: 95 °C for 3 min, 39 cycles of 95 °C for 10 s and 60 °C for 30 s using a SYBR Premix Ex TaqTM kit (Takara, Japan). The primers used were as follows: 5ʹ-GAGCTCAACCAGTATAGTGCC-3ʹ (forward), 5ʹ-ACATGATGATGTTGCCCCCAA-3ʹ (reverse) for circRNA_100146; 5ʹ-TCTGGCTCCGTGTCTTC-3ʹ (forward), 5ʹ-GTTGTGGTTGGTTGGTTTGT-3ʹ (reverse) for miR-149-5p; 5ʹ-CTCGCTTCGGCAGCACA-3ʹ (forward), 5ʹ-ACGCTTCACGAATTTGCGT-3ʹ (reverse) for U6; 5ʹ-TCAGTGGGGTTAGGTACATTC-3ʹ (forward), 5ʹ-TTTACAAGTAGACAGCGGTGA-3ʹ (reverse) for RNF2; 5ʹ-AAGGTCGGAGTCAACGGA-3ʹ (forward), 5ʹ-TTAAAAGCAGCCCTGGTGA-3ʹ (reverse) for GAPDH.
Cell Lines and Transfection
Normal bladder epithelial cells (HCV-29) and four bladder cancer cell lines (J82, 5637, SW780, and T24) were purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China). Cells were grown routinely in RPMI-1640 medium (Invitrogen, CA, USA) supplemented with 10% fetal bovine serum (Gibco, CA, USA) and cultured in a 37 °C humidified atmosphere containing 5% CO2. circRNA_100146 over-expression (circRNA) or knockdown (shRNA#1, shRNA#2, shRNA#3) plasmids, miR-149-5p mimic, RNF2 overexpression (RNF2) plasmid, and their control vector were obtained from GenePharma (Shanghai, China). J82 and T24 cells were seeded in 6-well plates 24 h prior to circRNA_100146 or miR-149-5p mimic or RNF2 transfection. When the cells were grown–70-80% confluence, transfection was carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol.
CCK8 Assay
The CCK-8 assay (Dojindo, Kumamoto, Japan) was used to determine the proliferation of J82 and T24 cells. In brief, 2000 cells per well were seeded into 96-well plates. After culturing for 0, 24, 48, 72, and 96 h, 10 μL of CCK-8 solution per well was added and incubated for 2 h. Then, the absorbance at 450 nm was determined using a SpectraMax iD5 microplate reader (Molecular Devices).
Colony Formation Assay
For the colony formation assay, 500 cells per well were seeded into 6-well plates. After culturing for 2 weeks, the clones were fixed and stained using crystal violet (Biolab, Shanghai, China). The colony number was counted under a microscope.
Cell Apoptosis
J82 and T24 cells were harvested and washed using ice-cold PBS, followed by staining with a Annexin V-FITC Apoptosis Detection Kit (JIANCHENG Biotech Co., Ltd., Shanghai, China). Cell apoptosis was analyzed using a flow cytometer (FACSCalibur, BD Biosciences). The cell apoptosis rate was analyzed using FlowJo 7.6.1 software.
Wound Healing and Invasion Assays
In the wound healing test, J82 and T24 cells were cultured in a 6-well plate until 100% confluence, and were then injured using the tip of a 20 mL pipette. Migration photos were taken at 0 and 48 h after the scratch. Invasive cells were detected via transwell invasion assay. Transwell filters were pre-coated with Matrigel (BD Biosciences). Approximately 5 × 104 cells were inoculated in serum-free medium in the upper chamber. The 1640 medium containing 20% FBS was added to the bottom chamber. After incubation for approximately 24 h, the invasive cells on the lower surface of the filter were fixed and examined under a microscope.
Western Blot
The protein was extracted using radioimmunoassay lysis buffer, and its concentration was determined using a BCA protein assay kit (HaiGene, Harbin, China). The protein was separated by 10% SDS-PAGE and Western blotting was performed using the following primary antibodies: anti-RNF2, at a dilution of 1:1000, (Cell Signaling); anti-GAPDH, at a dilution of 1:3000, (Proteintech). Membranes were then incubated with a peroxidase-conjugated secondary antibody, and specific bands were detected on an X-ray film (e-Blot, Shanghai, China) imaging system.
Immunohistochemistry
Tumor tissue or non-tumor tissue sections were deparaffinized with xylene, and then washed in serial dilutions of ethanol: (1) 100% ethanol, (2) 95% ethanol, (3) 80% ethanol, and finally in PBS. After washing, the endogenous peroxidase activity was quenched using methanol and H2O2 (0.3%) for 5 min. The slide was blocked in PBS containing 5% bovine serum albumin (BSA) for 30 min, and then incubated with RNF2 and Ki67 primary antibodies overnight at 4 °C (at a dilution of 1:100, Solaibao Biotechnology Co., Ltd., Beijing, China). After 24 h, the sections were incubated with the secondary antibody (1:1000; Solaibao Biotechnology) for 2 h.
Luciferase Reporter Assay
For the luciferase reporter assay, pmirGLO dual-luciferase vectors (Promega, Madison, WI, USA) were employed to construct dual-luciferase reporter plasmids. HEK-293T cells, which were purchased from the Chinese Academy of Sciences, were co-transfected with the corresponding plasmids and miRNA. Luciferase activity was determined using the dual-luciferase reporter kit (Promega). The relative activity of firefly luciferase was normalized to Renilla luciferase activity.
Tumor Xenograft in Nude Mice
The animal experiments were approved by the Ethics Committee of The Affiliated Huaian No.1 People’s Hospital of Nanjing Medical University. All the animal operations were performed in accordance with the Animal Policy and Welfare Committee of The Affiliated Huaian No.1 People’s Hospital of Nanjing Medical University. Briefly, 100 mL of T24 cells (1 × 106) transfected with shRNA-circRNA_100146 or scramble were subcutaneously injected into nude mice (n = 6 per group, 2 months of age). The tumor length and width were determined every 7 days, and the tumor volume was determined using the formula: tumor volume = πab2/6 (a = tumor length; b = tumor width).
Statistical Analysis
Student’s t-test and one-way ANOVA were used to assess statistical significance. GraphPad Prism 7 was used for statistical analysis. Survival rate was analyzed by Kaplan Meier curves and the Log rank test. The correlation between circRNA_100146 and clinicopathological features of bladder cancer patients was analyzed using the Pearson correlation test. P < 0.05 was considered statistically significant.
Results
The Expression of circRNA_100146 Was Abnormally Increased in Bladder Cancer Tissues and Cells
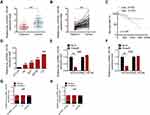
To explore the biological role of circRNA_100146 in bladder cancer, the expression of circRNA_100146 in bladder cancer clinical samples and cell lines was identified by qRT-PCR analysis. The results showed that the expression of circRNA_100146 was significantly increased in cancer tissues (n = 68) compared with that in adjacent non-cancerous tissues (n = 68) (Figure 1A and B). High expression of circRNA_100146 also displayed an unfavorable prognosis in bladder cancer patients (Figure 1C). In addition, the link between circRNA_100146 expression and the clinicopathological features of bladder cancer patients was studied. The results showed that high expression of circRNA_100146 was closely related to tumor stage, LN status, histological grade, and tumor size (Table 1). Furthermore, compared with the expression of circRNA_100146 in HCV-29 cells, circRNA_100146 was dramatically overexpressed in bladder cancer cell lines, including J82, 5637, SW780, and T24 (Figure 1D). The RNase R experiment was performed in J82 and T24 cells, and the data proved that circRNA_100146 was resistant to RNase digestion (Figure 1E and F). The expression of circRNA_100146 in the nucleus and cytoplasm was examined (Figure 1G and H). Results showed that circRNA_100146 was expressed both in the nucleus and in cytoplasm, while the expression abundance of circRNA_100146 was higher in the cytoplasm than in the nucleus.
CircRNA_100146 Regulated Cell Proliferation and Invasion of Bladder Cancer Cells
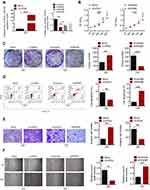
To determine whether circRNA_100146 affected bladder cell proliferation, apoptosis, invasion, or migration, J82 cells were treated with circRNA_100146 over-expression and T24 cells were transfected with shRNA-circRNA_100146 (shRNA). As shown in Figure 2A, circRNA_100146 was overexpressed in J82 cells, while circRNA_100146 was downregulated in T24 cells. After cell transfection, cell proliferation, clone-forming ability, apoptosis, invasion, and migration were evaluated by CCK8, crystal violet staining, Annexin V-FITC/PI staining, transwell invasion assay, and wound healing assay, respectively. Results showed that circRNA_100146 stimulated cell proliferation, invasion, and migration, and inhibited cell apoptosis in J82 cells; however, circRNA_100146 knockdown exerted anti-tumor effects on T24 cells (Figure 2B–F).
CircRNA_100146 and RNF2 Competitively Combined with miR-149-5p
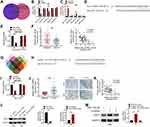
We then explored the molecular mechanism by which circRNA_100146 facilitated bladder cancer cell proliferation and invasion. The targeted miRNAs were predicted using the Circular RNA Interactome and StarBase. Five miRNAs, including miR-149-5p, miR-616-3p, miR-361-3p, miR-330-3p, and miR-526b-5p, were obtained (Figure 3A). The expression levels of the predicted miRNAs were detected in J82 or T24 cells. As displayed in Figure 3B, circRNA_100146 inhibited the expression of miR-149-5p and miR-361-3p, and silencing circRNA_100146 increased the expression of miR-149-5p and miR-361-3p (Figure 3C). circRNA_100146 transfection exhibited a greater regulatory impact on miR-149-5p expression. The predicted binding site of circRNA_100146 and miR-149-5p is shown in Figure 3D, which was further verified by dual-luciferase reporter (Figure 3E). Moreover, it was observed that the expression of miR-149-5p was downregulated in bladder cancer clinical samples and showed a negative correlation with the expression of circRNA_100146 (Figure 3F).
The targeted mRNAs of miR-149-5p were also predicted by TargetScan, miRDB, miRTarBase, and miRWalk. PDE1B, SLCO3A1, RNF2, AGO1, FASLG, and DCTN2 were screened (Figure 3G). RNF2 acts as an oncogene in several cancers, and was then selected for further experiments. Figure 3H shows that RNF2 could combine with miR-149-5p, and the dual-luciferase reporter also verified the combination of miR-149-5p and RNF2 (Figure 3I). Moreover, qRT-PCR and IHC analysis showed that the expression of RNF2 in bladder cancer tissues was upregulated (Figure 3J). Notably, RNF2 expression was negatively correlated with miR-149-5p expression in bladder cancer samples (Figure 3K). As evident from Western blot analysis, miR-149-5p negatively regulated the protein expression of RNF2 in J82 and T24 cells (Figure 3L). Additionally, circRNA_100146 overexpression promoted the expression of RNF2, while miR-149-5p mimic reversed the effect of circRNA_100146 in J82 cells (Figure 3M).
RNF2 Reversed the Effect of shRNA-circRNA_100146 and the in vivo Study
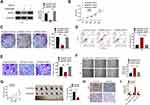
To further investigate the regulatory mechanism by which circRNA_100146 affects bladder cancer cells, T24 cells were transfected with Scramble+Vector, shRNA#1-circRNA_100146 (shRNA#1) +Vector, or shRNA#1+RNF2. The protein expression of RNF2 in T24 cells was determined (Figure 4A). As shown in Figure 4B–F, shRNA#1-circRNA_100146 transfection significantly suppressed the T24 cell proliferation, colony formation, invasion, and migration, and promoted cell apoptosis compared with the Scramble+Vector transfected cells. However, RNF2 overexpression remarkably reversed the effects of shRNA#1-circRNA_100146 transfection on T24 cells.
The biological effect of circRNA_100146 on bladder cancer tumor growth was evaluated using a xenograft mouse model. J82 cells transfected with Scramble or shRNA#1-circRNA_100146 were implanted subcutaneously into nude mice. Tumor growth was evaluated every 7 days. Knockdown of circRNA_100146 significantly delayed tumor growth in vivo, and decreased tumor size and weight (Figure 4G). Furthermore, downregulation of circRNA_100146 significantly decreased the expression levels of RNF2 and Ki67 (Figure 4H). The expression levels of circRNA_100146, miR-149-5p, and RNF2 in xenograft tumors were determined by qRT-PCR. Tumors derived from shRNA#1-circRNA_100146 transfected cells showed an obvious decrease in circRNA_100146 and RNF2 expression levels and exhibited a notable increase in miR-149-5p expression (Figure 4I).
Discussion
Nowadays, a great number of circRNAs are reported to regulate the occurrence and development of bladder cancer, such as circPICALM,17 circMTO1,11 and circPTK2.18 It was reported that circPICALM acted as an antioncogene that inhibited bladder cancer metastasis by sponging miR-1265.17 Xu et al indicated that circPTK2 accelerated the proliferation and migration of bladder cancer cells.18 A previous study indicated that circRNA_100146 was upregulated in bladder cancer, and circRNA_100146 acted as an oncogene in non-small cell lung cancer and thyroid cancer.13 In the present study, we explored the expression levels, biological role, and molecular mechanism of circRNA_100146 in bladder cancer. We first found that high expression of circRNA_100146 correlated with a poor prognosis in bladder cancer patients. Furthermore, correlation analysis revealed that elevated circRNA_100146 expression was observed in the bladder patients with LN+, high histological grade, and large tumor size. Moreover, it was detected that circRNA_100146 was overexpressed in bladder cancer cell lines compared to that in normal bladder epithelial cells. The results mentioned above revealed that circRNA_100146 might act as an oncogene in bladder cancer.
We subsequently determined the effects of circRNA_100146 overexpression or knockdown on bladder cancer cell proliferation, apoptosis, and invasion. It was observed that silencing circRNA_100146 expression significantly suppressed cell proliferation, invasion and migration, while promoted cell apoptosis of bladder cancer cells. Furthermore, upregulation of circRNA_100146 exerted a tumorigenic role in bladder cancer cells. Emerging evidence demonstrates that circRNA functions as an miRNA sponge, thus regulating a series of physiological activities. We next predicted the target miRNAs of circRNA_100146, including miR-149-5p, miR-616-3p, miR-361-3p, miR-330-3p, and miR-526b-5p. Our study confirmed that miR-149-5p expression can be regulated by circRNA_100146. Previous studies have reported that miR-149-5p is a tumor suppressor in several cancers, including gastric cancer,19 renal cell carcinoma,20 osteosarcoma,21 and nasopharyngeal carcinoma.22 Zhang et al reported that miR-149-5p was downregulated in gastric cancer, and was involved in the molecular mechanism of circNRIP1 regulating gastric cancer progression.19 Jin et al investigated that miR-149-5p was a tumor suppressor that inhibited cell invasion and promoted cell apoptosis in renal cell carcinoma.20 In this study, we reported that circRNA_100146 could combine with miR-149-5p. In addition, miR-149-5p was downregulated in bladder cancer samples, and the expression of miR-149-5p was negatively correlated with circRNA_100146 expression in clinical samples. The downstream mRNAs of miR-149-5p were predicted to be PDE1B, SLCO3A1, RNF2, AGO1, FASLG, and DCTN2. RNF2 was reported to act as an oncogene in gastric cancer,23 lung squamous cell carcinoma,24 mammary carcinoma,25 and urothelial carcinoma of the bladder.26 Li et al reported that over-expression of RNF2 played a crucial role in the prognosis of urothelial carcinoma of the bladder.26 In the present study, we found that circRNA_100146 sponged miR-149-5p and promoted RNF2 expression, thus accelerating cell proliferation and invasion of bladder cancer cells. As revealed by the clinical data analysis, RNF2 was over-expressed in bladder cancer and showed a positive correlation with circRNA_100146 expression. The rescue experiments showed that shRNA-circRNA_100146 inhibited the proliferation and invasion of T24 cells, while over-expression of RNF2 reversed the effects of shRNA-circRNA_100146 on T24 cells. These results indicated that circRNA_100146 competitively combined with miR-149-5p and increased RNF2 expression and promoted malignant phenotypes of bladder cancer cells. Finally, the tumor xenograft in nude mice experiments demonstrated that shRNA-circRNA_100146 could delay tumor growth, reduce tumor weight, and decrease tumor volume of bladder cancer.
In conclusion, the present study provides some evidence that circRNA_100146 is overexpressed in bladder cancer samples and that increased circRNA_100146 predicted a poor prognosis for bladder cancer patients. circRNA_100146 stimulated bladder cancer cell proliferation, migration, and invasion by sponging miR-149-5p and promoting RNF2 expression. There are also some limitations of our study. The downstream molecular mechanisms needs to be further explored. Only one cell line was performed in the rescue experiments and in vivo studies, more cell lines should be used for more studies. In addition, more efforts will be devoted to explore the effect and mechanism of circRNA_100146 on metastasis of bladder cancer. Our future work will cover these aspects. Our findings indicated that circRNA_100146 served as an oncogene in bladder cancer, and might be considered as a new biomarker for bladder cancer diagnosis and prognosis.
Disclosure
The authors report no conflicts of interest for this work. Co-first authors: Hengbing Wang and Xiaobing Niu.
References
1. Mathes J, Rausch S, Todenhofer T, et al. Trimodal therapy for muscle-invasive bladder cancer. Expert Rev Anticancer Ther. 2018;18:1219–1229. doi:10.1080/14737140.2018.1535314
2. Pham A, Ballas LK. Trimodality therapy for bladder cancer: modern management and future directions. Curr Opin Urol. 2019;29:210–215. doi:10.1097/MOU.0000000000000601
3. Alifrangis C, McGovern U, Freeman A, et al. Molecular and histopathology directed therapy for advanced bladder cancer. Nat Rev Urol. 2019;16:465–483. doi:10.1038/s41585-019-0208-0
4. Heide T, Maurer A, Eipel M, et al. Multiregion human bladder cancer sequencing reveals tumour evolution, bladder cancer phenotypes and implications for targeted therapy. J Pathol. 2019;248:230–242. doi:10.1002/path.5250
5. Wei Y, Zhang Y, Meng Q, et al. Hypoxia-induced circular RNA has_circRNA_403658 promotes bladder cancer cell growth through activation of LDHA. Am J Transl Res. 2019;11:6838–6849.
6. Yang X, Yuan W, Tao J, et al. Identification of circular RNA signature in bladder cancer. J Cancer. 2017;8:3456–3463. doi:10.7150/jca.19345
7. Okholm T, Nielsen MM, Hamilton MP, et al. Circular RNA expression is abundant and correlated to aggressiveness in early-stage bladder cancer. Npj Genom Med. 2017;2:36. doi:10.1038/s41525-017-0038-z
8. Liu T, Lu Q, Liu J, et al. Circular RNA FAM114A2 suppresses progression of bladder cancer via regulating NP63 by sponging miR-762. Cell Death Dis. 2020;11:47. doi:10.1038/s41419-020-2226-5
9. Su H, Tao T, Yang Z, et al. Circular RNA cTFRC acts as the sponge of MicroRNA-107 to promote bladder carcinoma progression. Mol Cancer. 2019;18:27. doi:10.1186/s12943-019-0951-0
10. He Q, Yan D, Dong W, et al. circRNA circFUT8 upregulates krupple-like factor 10 to Inhibit the metastasis of bladder cancer via sponging miR-570-3p. Mol Ther Oncolytics. 2020;16:172–187. doi:10.1016/j.omto.2019.12.014
11. Li Y, Wan B, Liu L, et al. Circular RNA circMTO1 suppresses bladder cancer metastasis by sponging miR-221 and inhibiting epithelial-to-mesenchymal transition. Biochem Biophys Res Commun. 2019;508:991–996. doi:10.1016/j.bbrc.2018.12.046
12. Liu F, Zhang H, Xie F, et al. Hsa_circ_0001361 promotes bladder cancer invasion and metastasis through miR-491-5p/MMP9 axis. Oncogene. 2020;39:1696–1709.
13. Chen L, Nan A, Zhang N, et al. Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol Cancer. 2019;18:13. doi:10.1186/s12943-019-0943-0
14. Xia F, Chen Y, Jiang B, et al. Hsa_circ_0011385 accelerates the progression of thyroid cancer by targeting miR-361-3p. Cancer Cell Int. 2020;20:49. doi:10.1186/s12935-020-1120-7
15. Huang W, Lu Y, Wang F, et al. Downregulation of circular RNA hsa_circ_0000144 inhibits bladder cancer progression via stimulating miR-217 and suppressing RUNX2 expression. Gene. 2018;678:337–342. doi:10.1016/j.gene.2018.08.036
16. Mao W, Huang X, Wang L, et al. Circular RNA hsa_circ_0068871 regulates FGFR3 expression and activates STAT3 by targeting miR-181a-5p to promote bladder cancer progression. J Exp Clin Cancer Res. 2019;38:169. doi:10.1186/s13046-019-1136-9
17. Yan D, Dong W, He Q, et al. Circular RNA circPICALM sponges miR-1265 to inhibit bladder cancer metastasis and influence FAK phosphorylation. Ebiomedicine. 2019;48:316–331. doi:10.1016/j.ebiom.2019.08.074
18. Xu ZQ, Yang MG, Liu HJ, et al. Circular RNA hsa_circ_0003221 (circPTK2) promotes the proliferation and migration of bladder cancer cells. J Cell Biochem. 2018;119:3317–3325. doi:10.1002/jcb.26492
19. Zhang X, Wang S, Wang H, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18:20. doi:10.1186/s12943-018-0935-5
20. Jin L, Li Y, Liu J, et al. Tumor suppressor miR-149-5p is associated with cellular migration, proliferation and apoptosis in renal cell carcinoma. Mol Med Rep. 2016;13:5386–5392. doi:10.3892/mmr.2016.5205
21. Xu RD, Feng F, Yu XS, et al. miR-149-5p inhibits cell growth by regulating TWEAK/Fn14/PI3K/AKT pathway and predicts favorable survival in human osteosarcoma. Int J Immunopathol Pharmacol. 2018;32:2058738418786656. doi:10.1177/2058738418786656
22. Kong YG, Cui M, Chen SM, et al. LncRNA-LINC00460 facilitates nasopharyngeal carcinoma tumorigenesis through sponging miR-149-5p to up-regulate IL6. Gene. 2018;639:77–84. doi:10.1016/j.gene.2017.10.006
23. Qi H, Xiao Z, Wang Y. Long non-coding RNA LINC00665 gastric cancer tumorigenesis by regulation miR-149-3p/RNF2 axis. Onco Targets Ther. 2019;12:6981–6990. doi:10.2147/OTT.S214588
24. Yang J, Yu F, Guan J, et al. Knockdown of RNF2 enhances the radiosensitivity of squamous cell carcinoma in lung. Biochem Cell Biol. 2019;97:589–599. doi:10.1139/bcb-2018-0252
25. Wu J, Wang H, Li Q, et al. The oncogenic impact of RNF2 on cell proliferation, invasion and migration through EMT on mammary carcinoma. Pathol Res Pract. 2019;215:152523. doi:10.1016/j.prp.2019.152523
26. Li XD, Chen SL, Dong P, et al. Overexpression of RNF2 is an independent predictor of outcome in patients with urothelial carcinoma of the bladder undergoing radical cystectomy. Sci Rep. 2016;6:20894. doi:10.1038/srep20894
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.