Back to Journals » Cancer Management and Research » Volume 13
Hsa_circRNA_000543 Predicts Poor Prognosis and Promotes Cervical Cancer Cell Progression Through Regulating miR-567/ZNF268 Axis
Authors Yao Z, Shu L, Yi Y, Qiao L
Received 15 January 2021
Accepted for publication 27 May 2021
Published 30 June 2021 Volume 2021:13 Pages 5211—5222
DOI https://doi.org/10.2147/CMAR.S302201
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Kenan Onel
Zhilan Yao,1,2 Liuping Shu,1,2 Yi Yi,1,2 Lifu Qiao1,2
1Department of Gynecology, Wujin Hospital Affiliated with Jiangsu University, Changzhou, Jiangsu Province, 213100, People’s Republic of China; 2Department of Gynecology, The Wujin Clinical College of Xuzhou Medical University, Changzhou, Jiangsu Province, 213100, People’s Republic of China
Correspondence: Yi Yi; Lifu Qiao
Department of Gynecology, Wujin Hospital Affiliated with Jiangsu University, No. 2 Yongning North Road, Changzhou, Jiangsu Province, 213100, People’s Republic of China
Tel +86-0519-85336190
Email [email protected]; [email protected]
Aim: Cervical cancer (CC) is the fourth most common cancer among women worldwide. We aimed to explore the role of hsa_circ_000543 played in CC.
Methods: The hsa_circ_000543 expressions in CC tissues and cells were measured by qRT-PCR. The correlation of hsa_circ_000543 expression and the clinical features of CC patients were analyzed by SPSS 20.0. The up- or down-regulated plasmids of hsa_circ_000543 were respectively transfected into CC cells. Cell proliferation, apoptosis and colony formation were detected through CCK-8 assay, flow cytometry and cell colony formation assay, respectively. The cell migration and invasion were evaluated by Transwell assay. The underlying molecular mechanism of hsa_circ_000543 was studied by bioinformatic prediction tools and luciferase reporter assay. Rescue experiments were performed to validate the regulation mechanism of hsa_circ_000543/miR-567/ZNF268 axis in CC.
Results: Hsa_circ_000543 was over-expressed in CC tissues and cells. The high expression of hsa_circ_000543 indicated poor prognosis of CC patients. Hsa_circ_000543 promoted cell proliferation, colony formation, migration and invasion, as well as inhibited cell apoptosis in CC cells. Hsa_circ_000543 directly targeted miR-567/ZNF268 in CC cell lines. In CC tumor tissues and cells, the hsa_circ_000543 expression was negatively correlated with miR-567 expression and showed a positive correlation with ZNF268 expression. The rescue experiments revealed that hsa_circ_000543 mediated cell proliferation, apoptosis, colony formation, migration and invasion of CC cells via regulating miR-567/ZNF268 axis.
Conclusion: Hsa_circ_000543 regulated CC cell activities through binding miR-567 and therefore enhancing ZNF268 expression.
Keywords: cervical cancer, circular RNA, hsa_circ_000543, miR-567, ZNF268
Introduction
Cervical cancer (CC) is the fourth most common female malignant tumor in the world, which seriously threatens the health of women all over the world. According to the Global Cancer Statistics 2018,1 there were about 569, 847 new patients with CC, and the number of deaths due to CC was as high as 311, 365. The incidence rate and mortality rate of CC vary greatly due to the geographical location and economic conditions of different regions.2 A study of 38 countries in five continents shows that the incidence rate of the economically scarce areas is increasing compared with the incidence rate of CC in the developed countries.3 In the past decades, despite the continuous improvement of vaccination and screening methods for human papilloma virus (HPV) vaccine, the incidence rate and mortality rate of CC are still increasing.4 Persistent high risk human papillomavirus (HR-HPV) infection is widely considered to be the cause of cervical intraepithelial neoplasia (CIN) and CC. However, only about 10% of women with persistent HR-HPV infection will develop into CC, which suggests that there may be other key factors involved in the process of CC.5
Circular RNA (circRNA) is a kind of noncoding RNA (ncRNA) that covalently binds to form a circular structure.6 CircRNA plays an important role in regulation of biological gene expression. With the emergence of new generation sequencing technology and the progress of bioinformatics, the regulatory role of circRNA in eukaryotic cells has been gradually recognized.7 CircRNA is a new non-coding RNA highly expressed in eukaryotic cells, which is not affected by RNA exonuclease and has evolutionary conservatism.8 The multiple biological functions of circRNA have become the focus of genetic research. Previous studies have shown that circRNA negatively regulates microRNA (miRNA) activity by competing for miRNA binding sites, which may act as miRNA sponge by regulating transcription, and play a crucial regulatory role in tumor development.8 There is increasing evidence that circRNAs are associated with a variety of human diseases, especially cancer.9 The main mechanism of circular RNA in cancer can be summarized as that circRNA acts as the sponge of miRNA, and indirectly regulates gene expression by specifically binding to miRNA.9
Hsa_circ_000543, also named as hsa_circ_0000326 (http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000326), is located at chr11:65272490–65272586. At present, there are few reports about the role of hsa_circ_000543 in tumor progression. Previous study reported that gene symbol of hsa_circ_000543 is MALAT1, and hsa_circ_000543 was abnormal expressed in lung adenocarcinoma.10 In addition, hsa_circ_000543 regulated lung adenocarcinoma cell activities by targeting miR338-3p/RAB14.10 However, the expression and functional role of hsa_circ_000543 in CC were unclear.
In the present study, we aimed to find the functional role and molecular mechanism of hsa_circ_000543 in CC. We found that hsa_circ_000543 was up-regulated in CC, and hsa_circ_000543 could modulate CC progression via regulating miR-567/ kruppel-associated box (KRAB) domain of zinc finger gene 268 (ZNF268) axis. Based on the data of the current study, targeting hsa_circ_000543 might be a novel strategy in CC therapy.
Materials and Methods
Patient Tissues
All CC patients involved in our study were pathologically diagnosed at the Wujin Hospital Affiliated with Jiangsu University from June 2013 to April 2015. A total of 50 pairs of CC tumor tissues and adjacent non-tumor tissues were obtained from the CC patients. All the CC patients had signed the written informed consent. Liquid nitrogen was used to place these tissues at once. These tissues were then stored in −80°C freezer. The current study was approved by the Ethics Committee of the Wujin Hospital Affiliated with Jiangsu University, and it was conducted in accordance with the Declaration of Helsinki. Then, the expressions of hsa_circ_000543 in the 50 cases of CC tumor tissues were detected, and the average value was calculated. The correlation between hsa_circ_000543 expression and CC clinical features [Age, International Federation of Gynecology and Obstetrics (FIGO) stage, Distant metastasis, Tumor size and Lymphatic metastasis] were subsequently analyzed.
Cell Lines
The immortalized human cervical epithelial cell line H8 and four human CC cell lines (Caski, ME180, C-33A and Siha) were all bought from BeNa Culture Collection (Suzhou, Jiangsu, China). Dulbecco’s Modified Eagle Medium (DMEM), purchased from Sigma (St Louis, MO, USA) was used to culture the H8, C-33A and Siha cell lines. Roswell Park Memorial Institute 1640 (RPMI 1640) medium, purchased from Thermo Fisher Scientific (Waltham, MA, USA), was applied to culture Caski cell lines. McCOY’s 5A medium, purchased from Thermo Fisher Scientific, was used to culture Me180 cell lines. 10% Fetal bovine serum (FBS), streptomycin (100 mg/mL) and penicillin (100 unit/mL), all purchased from Gibco (Waltham, MA, USA), were supplemented into the culture medium. And the cell lines used in the present study were all cultured in a incubator (containing 5% CO2) maintained at 37°C.
Cell Transfection
Hsa_circ_000543 overexpression plasmid (circRNA) and matched negative control plasmid (Vector), small interference RNA (siRNA) targeting hsa_circ_000543 (siRNA#1 and siRNA#2) and matched negative control (siNC), miR-567 overexpression plasmid (miR-567) and matched NC mimic, miR-567 down-regulation plasmid (miR-567 inhibitor) and matched NC inhibitor, siZNF268 and matched siControl were all designed and provided by GenePharma (Shanghai, China). These oligonucleotides (40 nM) and constructed vectors (600 ng) were transfected into CC cell lines (Caski and Siha) by Lipofectamine 3000 reagent that bought from Thermo Fisher Scientific for 24 h.
RNase R Treatment
RNase R, purchased from Epicentre (Madison, WI, USA), was used to treat the isolated RNA (10 μg) at the density of 3 U/mg for 1h at 37°C. And non-treatment was served as blank control. Quantitative real-time PCR (qRT-PCR) was subsequently carried out to assess the expression of hsa_circ_000543.
RNA Extraction and Quantitative Real-Time PCR
Total RNA was collected from CC tissues and cell lines using TRIzol which were purchased from TaKaRa (Shiga, Japan) with the accordance of manufacturer’s instructions. A PrimeScript RT Reagent Kit, provided by TaKaRa, was applied to reversely transcribe the RNA into cDNA. RNA expression was quantified by qRT-PCR, which was performed to quantify the RNA expression using SYBR Premix Ex Taq™ (TaKaRa). U6 was adapted as internal control for miRNA while GAPDH was adapted as internal control for circRNA and mRNA. The relative expression levels were measured by the 2−ΔΔCt method. The primers for hsa_circ_000543 are as follows: F: 5′-TTGAATAGATTTCAGCTTTATGC-3′, R: 5′-CCCATAACTGATCTGACTTTGT-3′. The primers for miR-338-3p are as follows: F: 5′-GGGTCCAGCATCAGTGATT-3′, R: 5′-GCGTTGTGTTGTGTTGTGTT-3′. The primers for miR-567 are as follows: F: 5′-AGGGAGTATGTTCTTCCAGGA-3′, R: 5′-GCGTTGTGTTGTGTTGTGTT-3′. The primers for miR-766 are as follows: F: 5′-GGAGGAGGAATTGGTGCT-3′, R: 5′-GTTGTGGTTGGTTGGTTTGT-3′. The primers for miR-935 are as follows: F: 5′-CCAGTTACCGCTTCCGC-3′, R: 5′-TCCTCCTCTCCTTCCTTCTC-3′. The primers for ZNF268 are as follows: F: 5′-CTCTATTCTGCTGCCCCTT-3′, R: 5′-TGTTTCGTTCTTGGAGAGGT-3′. The primers for GAPDH are as follows: F: 5′-CACCATTGGCAATGAGCGGTTC-3′, R: 5′-AGGTCTTTGCGGATGTCCACGT-3′. The primers for U6 are as follows: F: 5′-AGAGCCTGTGGTGTCCG-3′, R: 5′-CATCTTCAAAGCACTTCCCT-3′. All experiments were performed in triplicates.
Cell Counting Kit-8 (CCK-8) Assay
After transfection, CC cell lines were seeded into 96-well plates with a density of 5×103 cells/well. CCK-8 solution (10 μL), bought from Beyotime (Shanghai, China), was supplemented into every well for 2 h at the different time points (0, 24, 48 and 72 h) after transfection. Then, the absorbance was examined at 450 nm via a microplate reader (Bio-Rad, Hercules, CA, USA).
Cell Colony Formation Assay
After transfection, CC cell lines were seeded into 6-well plates with a density of 5×103 cells/well and then cultured for two weeks. Subsequently, the plates were fixed using paraformaldehyde (4%). A crystal violet solution (0.5%) was applied to stain the colonies and the number of colony cells was counted.
Flow Cytometry
Cells apoptosis was evaluated by FITC Annexin V Apoptosis Detection Kit (BD Pharmingen, San Diego, CA, USA) using flow cytometry that provided by Becton Dickinson (San Jose, CA, USA) according to the manufacturer’s protocols. After transfection for 24 h, Caski and Siha cells were collected and stained using FITC Annexin V and propidium iodide (PI) at the density of 1×106 cells. Then, the stained Caski and Siha cells were assessed using flow cytometry. CellQuest software, provided by BD Bioscience, was subsequently performed to analyze the data.
Cell Migration and Invasion
The migration and invasion of Caski and Siha cells were examined by a Transwell chamber coated with or without Matrigel (Solarbio, Beijing, China). The experiments were performed followed by the manufacturer’s instructions. After the Caski and Siha cells were treated with transfection for 24 h, the migration or invasion cells were fixed at the bottom with 4% polyoxymethylene (Ybscience, Shanghai, China). Then, the Caski and Siha cells were dyed with 0.1% crystal violet (Solarbio). The stained Caski and Siha cells were counted with a microscope and photographed.
Bioinformatics Analysis
In the current study, the potential targets of circRNA were predicted using Circular RNA Interactome (https://circinteractome.nia.nih.gov/), the potential targets of miRNA were predicted using several bioinformatics tools, including TargetScan (http://www.targetscan.org/vert_71/), miRDB (http://mirdb.org/), miRTarBase (http://mirtarbase.cuhk.edu.cn/) and miRWalk (http://mirwalk.umm.uni-heidelberg.de/).
Luciferase Reporter Assays
The circ-000543 wild sequence, ZNF268-3′UTR wild sequence and their mutations with or without miR-567 binding sites were synthesized and then subcloned into luciferase reporter vectors psiCHECK-2, which was bought from Promega (Madison, WI, USA). HEK293T cells were implanted in 24-well plates at the density of 3×104 cells/well in triplicate. Then, miR-567 or NC mimic were cotransfected with corresponding psiCHECK-2 plasmids. After co-transfection for 48 h, dual-luciferase reporter assay system that provided by Promega was proceeded to conduct the luciferase reporter assays according to the manufacturer’s instructions. Relative luciferase activity was subsequently normalized to the Renilla luciferase internal control.
Western Blot
Total protein in the present study was isolated using RIPA lysis buffer, which was purchased from Thermo Fisher Scientific and quantified with a bicinchoninic acid (BCA) kit purchased from Sigma-Aldrich. Protein lysates were exposed to SDS-PAGE gels. And, the protein samples were then electrotransferred onto a polyvinylidene fluoride (PVDF) membrane that bought from Solarbio (Beijing, China). The membranes were subsequently blocked in 5% skimmed milk and incubated with primaries (Abcam, Cambridge, MA, USA) and horseradish peroxidase-conjugated secondary antibody (1:1000 dilution, Cell Signaling Technology, USA). At last, the bands were incubated with an enhanced chemiluminescence kit (ECL, Solarbio). GAPDH was regarded as a internal control. Primary antibodies involved in our study were used as follows: anti-GAPDH (1:5000 dilution, ab70699) and anti-ZNF268 (1:1000 dilution, orb647833, LifeSpan BioSciences).
Statistical Analysis
All the experiments carried out in our study were repeated for 3 times. Statistical analysis was performed with GraphPad Prism 7.0 (GraphPad Software Inc., CA, USA) and SPSS 20.0 software (SPSS, Chicago, IL, USA). Data were shown as means ± SD. Statistical significance was analyzed by one-way ANOVA, Student’s t-test or Pearson’s χ2 test. Kaplan–Meier method was applied to calculate the overall survival rates, and the Log rank test was used. Differences were deemed significant at P<0.05.
Results
Hsa_circ_000543 Was Abnormally Expressed in CC Tumor Tissues and Cell Lines
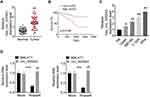
We firstly collected 50 pairs of CC tumor tissues and matched non-tumor tissues, and qRT-PCR was performed to detect the hsa_circ_000543 expression. As shown in Figure 1A, the expression of hsa_circ_000543 in Tumor group was significantly higher than that of Normal group (P<0.01). Then, the 50 cases of CC tumor tissues were divided into hsa_circ_000543 high expression group (n=25) and hsa_circ_000543 low expression group (n=25), and the relationship of hsa_circ_000543 expression level and overall survival of CC patients was analyzed. Obviously, the relative high hsa_circ_000543 expression indicated shorter survival time, while the relative low hsa_circ_000543 expression was related to longer survival time (P=0.0189, Figure 1B). The expression level of hsa_circ_000543 in CC cell lines was also assessed. Data from qRT-PCR exhibited that hsa_circ_000543 was markedly over-expressed in human CC cell lines including Caski, ME180, C-33A and Siha when compared with that in H8 cell line (P<0.05, Figure 1C). Besides, Figure 1C also showed that hsa_circ_000543 expressed highest in Siha cell lines and lowest in Caski cell lines among the four experimental cell lines in our study. Therefore, Caski and Siha cell lines were selected to carry out for the following experiments. In addition, hsa_circ_000543 was more resistant to RNase R digestion in Siha and Caski cell lines compared with linear MALAT1 mRNA (P<0.01, Figure 1D). Moreover, we also found that hsa_circ_000543 expression was remarkably associated with FIGO stage and tumor size of CC patients via χ2 test (Table 1). These data illustrated that hsa_circ_000543 was up-regulated in CC clinical samples and cell lines, and high expression of hsa_circ_000543 predicted a poor prognosis of CC patients.
 |
Table 1 Relationship Between Hsa_circ_000543 Expression and Clinical Features of Cervical Cancer Patients |
Hsa_circ_000543 Regulated Cell Proliferation, Colony Formation and Apoptosis in CC
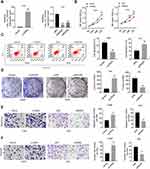
Caski cells were transfected with circRNA to over express hsa_circ_000543, while Siha cells were transfected with two siRNAs (siRNA#1 and siRNA#2) to knock down hsa_circ_000543 expression. The transfection efficiency was detected using qRT-PCR. As shown in Figure 2A, the expression of hsa_circ_000543 was enhanced in Caski cells, while both siRNA#1 and siRNA#2 reduced the hsa_circ_000543 expression in Siha cells (P<0.01). The siRNA#1 showed higher knockdown efficiency compared to siRNA#2; thus, siRNA#1 was selected to perform further experiments. CCK-8 assay presented that hsa_circ_000543 up-regulation markedly promoted cell proliferation in Caski cells, whereas hsa_circ_000543 knockdown effectively inhibited cell proliferation in Siha cells (P<0.01, Figure 2B). Data from flow cytometry displayed that circRNA transfection significantly reduced cell apoptosis in Caski cells, while siRNA#1 transfection apparently facilitated cell apoptosis in Siha cells (P<0.01, Figure 2C). In addition, colony formation assay suggested that the up-regulation of hsa_circ_000543 accelerated cell colony formation in Caski cells and the down-regulation of hsa_circ_000543 showed the opposite effect on cell colony formation of Siha cells (P<0.01, Figure 2D). Cell migration and invasion were examined by Transwell assay, and hsa_circ_000543 up-regulation boosted cell migration and invasion in Caski cells, whereas hsa_circ_000543 knockdown inhibited cell migration and invasion in Siha cells (P<0.01, Figure 2E and F). These functional experiments elucidated that hsa_circ_000543 stimulated cell proliferation and colony formation and suppressed cell apoptosis in CC cell lines.
Hsa_circ_000543 Directly Bound with miR-567
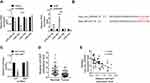
CircRNAs could serve as inhibitors of their interacting microRNAs.11 Therefore, miR-338-3p, miR-567, miR-766 and miR-935 were predicted to be interacting miRNAs of hsa_circ_000543 by using online bioinformatics tools. The expressions of miR-338-3p, miR-567, miR-766 and miR-935 in CC cell lines that with hsa_circ_000543 up- or down-regulation were measured using qRT-PCR. As shown in Figure 3A, the miR-338-3p and miR-567 expressions were negatively regulated by hsa_circ_000543 transfection (P<0.05). Hsa_circ_000543 showed the most obvious regulatory role on miR-567 expression in Caski and Siha cell lines (Figure 3A); therefore, we assumed that miR-567 was the potential targeted miRNA of hsa_circ_000543 in CC cells. The binding sites of hsa_circ_000543 and miR-567 are displayed in Figure 3B. Luciferase reporter assay exhibited that miR-567 significantly reduced the relative luciferase activity of hsa_circ_000543-WT vectors in contrast to the NC mimic (P<0.01), while hsa_circ_000543-MUT vectors did not change (Figure 3C). In addition, the expression of miR-567 in the 50 pairs of tumor and normal tissue samples were evaluated by qRT-PCR. Data of qRT-PCR presented that miR-567 was low-expressed in CC tumor tissues when compared to that in non-tumor tissues (P<0.01, Figure 3D). Moreover, Pearson’s χ2 test suggested that the expression of hsa_circ_000543 was negatively related to the expression of miR-567 in the 50 cases of CC clinical samples (P=0.0002, Figure 3E). These data demonstrated that hsa_circ_000543 acted as a sponge of miR-567 in CC cells.
MiR-567 Directly Targeted ZNF268
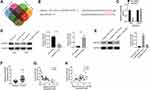
ZNF268 was predicted as the downstream functional gene of miR-567 via TargetScan, miRDB, miRTarBase and miRWalk (Figure 4A), and the binding sites of ZNF268 and miR-567 are shown in Figure 4B. Luciferase reporter assay displayed that miR-567 significantly inhibited the relative luciferase activity of ZNF268-3ʹUTR-WT vectors when compared to NC mimic (P<0.01), while the relative luciferase activity of ZNF268-3ʹUTR-MUT did not change (Figure 4C). The expressions of ZNF268 in CC cell lines that with miR-567 mimic or inhibitor transfection were identified using Western blot and qRT-PCR. As shown in Figure 4D, the miR-567 mimic obviously decreased the ZNF268 expression in Caski cells, while the knockdown of miR-567 evidently enhanced the ZNF268 expression in Siha cell lines (P<0.01). The Figure 4E exhibited that the expression of ZNF268 was increased by hsa_circ_000543 overexpression transfection, and it was remarkably reduced when miR-567 mimic was cotransfected (P<0.01, Figure 4E). In addition, the expression of ZNF268 in the 50 pairs of tumor and normal tissue samples was tested using qRT-PCR. The results presented that ZNF268 was markedly over-expressed in CC tumor tissues when compared to that in non-tumor tissues (P<0.01, Figure 4F). Moreover, Pearson’s χ2 test suggested that the expression of miR-567 was negatively related to the expression of ZNF268 in the CC tumor tissues (P<0.01, Figure 4G). The expression of hsa_circ_000543 was positively associated with expression of ZNF268 (P=0.0052, Figure 4H). These results suggested that ZNF268 was the downstream mRNA of miR-567 in CC cells.
Hsa_circ_000543 Affected CC Cell Activities via Modulating miR-567/ZNF268 Axis
To examine the molecular mechanism by which hsa_circ_000543 affected CC cell activities, we performed rescue experiment. The Caski cells were divided into three groups (Vector + siControl, circRNA + siControl and circRNA + siZNF268) based on different co-transfections. The protein expression of ZNF268 was examined by Western blot. The ZNF268 expression was increased by hsa_circ_000543 overexpression transfection, while siZNF268 transfection effectively reversed the increase of ZNF268 expression in Caski cells (P<0.01, Figure 5A). As shown in Figure 5B, the knockdown of ZNF268 obviously recovered the influences of hsa_circ_000543 over expression worked on CC cell proliferation (P<0.01). Similarly, ZNF268 knockdown also remarkably reduced the cell colony formation increases (P<0.01, Figure 5C) and enhanced cell apoptosis decreases (P<0.01, Figure 5D), which were both induced by the up-regulation of hsa_circ_000543. In summary, these data proved that hsa_circ_000543 accelerated cell proliferation and colony formation and repressed cell apoptosis in CC cell lines through regulating miR-567/ZNF268 axis. The result of Transwell assay showed that hsa_circ_000543 promoted cell migration and invasion in Caski cells, while siZNF268 decreased cell migration and invasion induced by hsa_circ_000543 overexpression (P<0.01, Figure 5E).
Discussion
With the improvement of screening methods, the incidence rate of CC has been reduced in the developed countries.12 However, in developing countries, CC is still the main cause of death for women due to the lack of universal screening methods. HPV infection, cellular, immune, epigenetic or environmental factors have impacts on the occurrence and development of CC.13 Previous studies have shown that HPV can be detected in more than 99% of CC patients, but HPV infection is not a sufficient factor for CC.14 Therefore, it is of great clinical significance to explore the pathogenesis of CC and analyze the role of small molecules in the CC progression. CircRNA is a kind of endogenous ncRNA, which is in a closed loop and widely exists in various organisms.6 The most significant function of circRNA is that it can be used as a miRNA sponge to compete for miRNA binding sites and regulate mRNA expression.8 The abnormal expression of circRNAs leads to the disorder of gene expression regulation network, which can activate the CC pathway abnormally and induce the growth of CC cells.15 More and more studies have shown that circRNAs are involved in the regulation of signal transduction pathway and gene expression related to CC, which may become a new biomarker or a potential target of radiotherapy for CC.
At present, there are several studies on the role of circRNAs in CC, such as the basic research on the mechanism of action in CC. Song et al.16 reported that hsa_circRNA_101996 enhanced CC cell invasion and proliferation via modulating miR-8075/TPX2 expressions. Zhang et al.17 found that hsa_circ_0000069, located at chr1:47745912–47748131, was over-expressed in CC tissues and cell lines, and it promoted CC cell activities through sponging miR-873-5p to accelerate TUSC3 expression in vivo and in vitro. Ou et al.18 proved that circ-AKT1, induced by TGF-β, acted as an oncogene in CC by inhibiting miR-924 to increase the expression of AKT1 in vivo and in vitro. In this study, we focus on hsa_circ_000543, which was found to be an oncogene in lung adenocarcinoma.10 We firstly explored the functional role of hsa_circ_000543 acted in CC.
We proved that hsa_circ_000543 was highly expressed in CC tumor tissues and cell lines, and hsa_circ_000543 expression was associated with overall survival rate, tumor size and FIGO stages of CC patients. The overexpression of hsa_circ_000543 indicated poor prognosis of CC patients. Then, we found that hsa_circ_000543 knockdown inhibited cell proliferation and colony formation and promoted cell apoptosis in human CC cell lines Siha, while hsa_circ_000543 up-regulation accelerated cell proliferation and colony formation and repressed cell apoptosis in human CC cell lines Caski. These findings suggested that hsa_circ_000543 might act as an oncogene in CC.
CirRNAs could serve as inhibitors of their interacting miroRNAs.11 Subsequently, the target miRNA of hsa_circ_000543 in CC were predicted and verified. Previous studies reported that miR-567 could affect progression of various tumors, such as breast cancer,19 gastric cancer,20 lung adenocarcinoma21 and osteosarcoma.22 Han et al. reported that miR-567 decreased the trastuzumab resistance of breast cancer cells, which was by inhibiting the expression of ATG5.19 MiR-567 acted as a tumor suppressor and regulated chemoresistance in gastric cancer by regulating PIK3AP1-PI3K/AKT-c-Myc pathway.20 MiR-567 also inhibited cell proliferation, migration and invasion by inhibiting the FGF5 expression in osteosarcoma.22 However, the role of miR-567 in CC has not been reported. In the current study, we investigated that hsa_circ_000543 sponged to miR-567 in CC. The expression of miR-567 was decreased in Caski cells with hsa_circ_000543 overexpression, and increased in Siha cells with hsa_circ_000543 knockdown. Besides, the expression of miR-567 in CC tumor tissue samples was lower than that in adjacent non-tumor tissue samples. And, in the CC tumor tissues, hsa_circ_000543 expression was proved to be negatively associated with miR-567 expression. ZNF268, a KRAB-containing zinc finger protein, includes the KRAB and zinc finger domains and acts as a transcriptional suppressor.23 It was reported to have effects on several cancers, such as hematological malignancy,24 ovarian cancer25 and leukemia.26 Especially, ZNF268 was proved to be up-regulated in CC and promote CC progression via activating NF-κB signaling pathway.27 Therefore, we predicted and validated that ZNF268 was the downstream mRNA of miR-567 in CC. We confirmed that the expression of ZNF268 was increased in CC cell lines when the expression of hsa_circ_000543 was up-regulated or the expression of miR-567 was down-regulated, while the expression of ZNF268 was inhibited in CC cells when the expression of miR-567 was up-regulated. Besides, ZNF268 expression in the CC tumor tissue samples was significantly higher than that in normal tissues. In addition, in the CC tumor tissues, the expression of ZNF268 was negatively related to the expression of miR-567 and positively associated with the expression of hsa_circ_000543. Finally, we demonstrated that hsa_circ_000543 promoted CC cell proliferation, colony formation, migration and invasion, and reduced cell apoptosis by restricting miR-567 expression to enhance ZNF268 expression according to the rescue experiments. There are some limitations of our current study. First, we chose Siha cell lines (hsa_circ_000543 expressed highest) to knock down hsa_circ_000543, and chose Caski cell lines (hsa_circ_000543 expressed lowest) to over express hsa_circ_000543. One more cell line should be used to validate the results, respectively. And, the thorough mechanism of hsa_circ_000543 on CC and its effect on CC in vivo need to be further studied. Our future work plans will cover these studies.
In conclusion, the current study proved that hsa_circ_000543 was over-expressed in CC clinical samples and displayed a poor prognosis. Hsa_circ_000543 contributed to cell proliferation and colony formation and suppressed cell apoptosis in CC cells via sponging miR-567 to enhance the ZNF268 expression. Our results suggest that hsa_circ_000543/miR-567/ZNF268 axis plays an important role in CC progression, and it might be a novel biomarker for CC therapy.
Disclosure
The authors declare no conflict of interest.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
2. Arbyn M, Weiderpass E, Bruni L, et al. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Global Health. 2020;8(2):e191–e203. doi:10.1016/S2214-109X(19)30482-6
3. Vaccarella S, Lortet-Tieulent J, Plummer M, et al. Worldwide trends in cervical cancer incidence: impact of screening against changes in disease risk factors. Eur J Cancer. 2013;49(15):3262–3273. doi:10.1016/j.ejca.2013.04.024
4. Small W, Bacon MA, Bajaj A, et al. Cervical cancer: a global health crisis. Cancer. 2017;123(13):2404–2412. doi:10.1002/cncr.30667
5. Crow JM. HPV: the global burden. Nature. 2012;488(7413):S2–S3. doi:10.1038/488S2a
6. Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–881. e13. doi:10.1016/j.cell.2018.12.021
7. Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol. 2017;14(8):1035–1045. doi:10.1080/15476286.2016.1271524
8. Hsiao K-Y, Sun HS, Tsai S-J. Circular RNA–new member of noncoding RNA with novel functions. Exp Biol Med. 2017;242(11):1136–1141. doi:10.1177/1535370217708978
9. Zhang Z, Xie Q, He D, et al. Circular RNA: new star, new hope in cancer. BMC Cancer. 2018;18(1):1–10. doi:10.1186/s12885-018-4689-7
10. Xu Y, Yu J, Huang Z, et al. Circular RNA hsa_circ_0000326 acts as a miR-338-3p sponge to facilitate lung adenocarcinoma progression. J Exp Clin Cancer Res. 2020;39(1):1–14. doi:10.1186/s13046-020-01556-4
11. Zhu K-P, Zhang C-L, Ma X-L, et al. Analyzing the interactions of mRNAs and ncRNAs to predict competing endogenous RNA networks in osteosarcoma chemo-resistance. Mol Therapy. 2019;27(3):518–530. doi:10.1016/j.ymthe.2019.01.001
12. Curry SJ, Krist AH, Owens DK, et al. Screening for cervical cancer: US Preventive Services Task Force recommendation statement. JAMA. 2018;320(7):674–686. doi:10.1001/jama.2018.10897
13. Vu M, Yu J, Awolude OA, et al. Cervical cancer worldwide. Curr Probl Cancer. 2018;42(5):457–465. doi:10.1016/j.currproblcancer.2018.06.003
14. Kessler TA. Cervical Cancer: Prevention and Early Detection. In Seminars in Oncology Nursing. Elsevier; 2017.
15. Chaichian S, Shafabakhsh R, Mirhashemi SM, et al. Circular RNAs: a novel biomarker for cervical cancer. J Cell Physiol. 2020;235(2):718–724. doi:10.1002/jcp.29009
16. Song T, Xu A, Zhang Z, et al. CircRNA hsa_circRNA_101996 increases cervical cancer proliferation and invasion through activating TPX2 expression by restraining miR‐8075. J Cell Physiol. 2019;234(8):14296–14305. doi:10.1002/jcp.28128
17. Zhang S, Chen Z, Sun J, et al. CircRNA hsa_circRNA_0000069 promotes the proliferation, migration and invasion of cervical cancer through miR-873-5p/TUSC3 axis. Cancer Cell Int. 2020;20(1):1–12. doi:10.1186/s12935-019-1086-5
18. Ou R, Mo L, Tang H, et al. circRNA-AKT1 Sequesters miR-942-5p to Upregulate AKT1 and Promote Cervical Cancer Progression. Mol Therapy Nucleic Acids. 2020;20:308–322. doi:10.1016/j.omtn.2020.01.003
19. Han M, Hu J, Lu P, et al. Exosome-transmitted miR-567 reverses trastuzumab resistance by inhibiting ATG5 in breast cancer. Cell Death Dis. 2020;11(1):1–15. doi:10.1038/s41419-020-2250-5
20. Zhang F, Li K, Yao X, et al. A miR-567-PIK3AP1-PI3K/AKT-c-Myc feedback loop regulates tumour growth and chemoresistance in gastric cancer. EBioMedicine. 2019;44:311–321. doi:10.1016/j.ebiom.2019.05.003
21. Yu C, Tian F, Liu J, et al. Circular RNA cMras inhibits lung adenocarcinoma progression via modulating miR‐567/PTPRG regulatory pathway. Cell Prolif. 2019;52(3):e12610. doi:10.1111/cpr.12610
22. Liu D, Zhang C, Li X, et al. MicroRNA-567 inhibits cell proliferation, migration and invasion by targeting FGF5 in osteosarcoma. EXCLI j. 2018;17:102. doi:10.17179/excli2017-932
23. Sun Y, Gou D, Liu H, et al. The KRAB domain of zinc finger gene ZNF268: a potential transcriptional repressor. IUBMB Life. 2003;55(3):127–131. doi:10.1080/1521654031000110208
24. Zhao Z, Wang D, Zhu C, et al. Aberrant alternative splicing of human zinc finger gene ZNF268 in human hematological malignancy. Oncol Rep. 2008;20(5):1243–1248.
25. Hu L, Wang W, Cai J, et al. Aberrant expression of ZNF268 alters the growth and migration of ovarian cancer cells. Oncol Lett. 2013;6(1):49–54. doi:10.3892/ol.2013.1318
26. Wang D, Guo M-X, Hu H-M, et al. Human T-cell leukemia virus type 1 oncoprotein tax represses ZNF268 expression through the cAMP-responsive element-binding protein/activating transcription factor pathway. J Biol Chem. 2008;283(24):16299–16308. doi:10.1074/jbc.M706426200
27. Wang W, Guo M, Hu L, et al. The zinc finger protein ZNF268 is overexpressed in human cervical cancer and contributes to tumorigenesis via enhancing NF-κB signaling. J Biol Chem. 2012;287(51):42856–42866. doi:10.1074/jbc.M112.399923
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.