Back to Journals » OncoTargets and Therapy » Volume 13
Hsa_circ_103973 Acts as a Sponge of miR-335 to Promote Cervical Cancer Progression
Authors Zhu Y, Jiang X, Zhang S , Wang L , Zhou Q, Jiang J
Received 14 May 2019
Accepted for publication 20 August 2019
Published 28 February 2020 Volume 2020:13 Pages 1777—1786
DOI https://doi.org/10.2147/OTT.S215736
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sanjeev K. Srivastava
Yingping Zhu, 1 Xuelu Jiang, 1 Shuo Zhang, 2 Lingcong Wang, 3 Qun Zhou, 1 Jun Jiang 1
1Department of Obstetrics and Gynecology, The First Affiliated Hospital of Zhejiang University of Traditional Chinese Medicine, Hangzhou 310006, People’s Republic of China; 2Department of Gastroenterology, The First Affiliated Hospital of Zhejiang University of Traditional Chinese Medicine, Hangzhou 310006, People’s Republic of China; 3Department of Critical Care Medicine, The First Affiliated Hospital of Zhejiang University of Traditional Chinese Medicine, Hangzhou 310006, People’s Republic of China
Correspondence: Jun Jiang; Yingping Zhu
Department of Obstetrics and Gynecology, The First Affiliated Hospital of Zhejiang University of Traditional Chinese Medicine, No. 54, Youdian Road, Shangcheng District, Hangzhou 310006, People’s Republic of China
Tel/Fax +86 571-86620269
Email [email protected]; [email protected]
Background: Cervical cancer (CC) ranks as the second most common malignancy in women, accounting for more two 2 million deaths every year in the world. Recently, circular RNAs (circRNAs) have been reported to regulate the progression of multiple human tumors; however, whether it involves in CC remains largely elusive.
Materials and Methods: Two GEO circRNA expression profiles (GSE102686, GSE113696) were downloaded to analyze the differentially expressed circRNAs using bioinformatics methods. Expression of circ_103973, miR-335 and PPP6C in CC tissues and cell lines were examined by qRT-PCR. Cell apoptosis was assessed with PI/Annexin-V double staining followed by the analysis of flow cytometry. Cell proliferation was evaluated by MTT and colony formation assays. Interaction between circ_103973 and miR-335, as well as miR-335 and PPP6C, were verified by dual-luciferase reporter assay.
Results: Circ_103973 was found to be highly expressed in both GSE102686 and GSE113696 datasets as well as in CC tissue samples and cell lines. Higher levels of circ_103973 were correlated to a worse outcome of CC patients. Knockdown of circ_103973 significantly promoted CC cell apoptosis and inhibited CC cell proliferation in vitro. Mechanistically, we demonstrated that circ_103973 served as a sponge of miR-335, which directly targeted PPP6C in CC cells. miR-335 was found to be decreased in CC, while PPP6C was found to be increased in CC. Moreover, anti-miR-335 could reverse the inhibitory effects of circ_103973 knockdown on CC cell proliferation, and this phenomenon could be blocked by si-PPP6C.
Conclusion: Circ_103973 promoted CC cell proliferation in vitro by physically binding miR-335, which further targeted and regulated PPP6C.
Keywords: cervical cancer (CC), circ_103973, miR-335, PPP6C, proliferation, apoptosis
Introduction
Cervical cancer (CC) ranks as the second most common malignancy in women, accounting for approximately 500,000 new cases and more than 250,000 deaths every year in the world.1,2 Estimation from the American Cancer Society indicates that 13,170 new cases will be diagnosed in the United State in 2019, and more than a third of patients will die.3 Although the exact pathogenesis of CC remains largely unclear, increasing evidences have shown that the persistent infection of high-risk kind human papillomavirus (HR-HPV) might be one of the critical pathogenic factors in its initiation and progression.4 Recent years, with the development and extensive application of HPV vaccine, the incidence of CC has been reduced to a considerable degree, making the morbidity and mortality of CC under effective control.5 However, despite the tremendous advancement in medical technologies in the past decades, the molecular mechanisms of CC remain largely unknown. Therefore, it is urgent to understand more about the molecular mechanisms of CC and identify several novel critical agents for the diagnosis and therapy of CC.
Circular RNAs (circRNAs) are a newly identified type of non-coding RNAs in mammal cells with a circular structure.6 Unlike linear RNAs, circRNAs have no 5ʹ-cap and 3ʹ-poly A tail, which enable circRNAs resistant to the degradation of exonuclease and RNase R.7 Due to this character, circRNAs have been considered to be more conservative and stable compared to multiple linear RNAs, such as micro RNAs (miRNAs) and long non-coding RNAs (lncRNAs).8,9 Moreover, circRNAs' expression usually exhibited cell-, tissue- and even time-specific manner.8 Thus, circRNAs were considered to be an important type of diagnostic and therapeutic agent of human tumors.10 During the past decades, circRNAs have demonstrated to play a critical role in the initiation and progression of multiple human cancers, including breast cancer, lung cancer and liver cancer.11 The implication of circRNAs in the pathogenesis of CC is supported by the microarray analysis showing the expression profiles of circRNAs are dysregulated in CC tissues compared to normal controls.12,13 The dysregulation of circRNAs expression profile in CC tissues suggested that they might play a critical role in the tumorigenesis of CC. However, the roles of most of circRNAs in the pathogenesis of CC remain undetermined.
In the present study, we aim to identify specific circRNAs that correlated to the tumorigenesis of CC, attempting to explore several novel diagnostic and therapeutic targets for CC patients. Bioinformatics analysis was performed in two GSE datasets (GSE102686 and GSE113696) to identify the dysregulated expressed circRNAs of CC tissue samples. As results showed that circ_103973 was identified to be upregulated in both GSE102686 and GSE113696 datasets. qRT-PCR detection of circ_103973 confirmed the upregulation of circ_103973 in CC tissue samples and cell lines. In the following functional assays, we found that circ_103973 might act as an oncogene of CC. In mechanism, circ_103973 was demonstrated to sever as a sponge of miR-335, which further targeted and regulated the expression of PPP6C. In conclusion, circ_103973 promoted CC cell proliferation in vitro by indirectly regulating the expression of PPP6C via sponging miR-335.
Materials and Methods
CC Tissue Samples
CC and adjacent normal tissue samples (5 cm beyond the boundary of the cervical cancer tissues) were collected from 23 patients who underwent surgery at the First Affiliated Hospital of Zhejiang University of Traditional Chinese Medicine during 2015–2019. Patients were an average of 59 years old, with a range of 29–78 years. The inclusion criteria included patients with pathologically diagnosed cervical cancer; patients are willing to provide written informed consents. The exclusion criteria included patients who have received chemotherapy and radiotherapy; patients with cognitive and communication disorders; patients who do not cooperate with the examination; patients with other severe visceral diseases or complications. All patients signed an informed consent document for diagnosis and research on tissue specimens before being enrolled in the project. All subjects gave written informed consent in accordance with the Declaration of Helsinki principles. All tissue samples were dipped in liquid nitrogen immediately removed from patients and then stored at −80°C.
Cell Lines
The human cervical epithelial cell line (HCerEpiC), and four CC cell lines (HeLa, CaSki, C33A, SiHa) were provided by the Committee on Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). All types of cell lines were cultured in the Dulbecco’s Modified Eagle’s Medium (DMEM; Invitrogen, Waltham, MA) containing 10% fetal bovine serum, 100 U/mL of penicillin, 100 μg/mL of streptomycin (Sigma-Aldrich) under 5% CO2, 95% O2 and 37°C.
Cell Transfection
MiR-335 mimics, anti-miR-335 and corresponding scrambles were purchased from Genepharma Co., Ltd. (Shanghai, China). HeLa and C33A cells (5×104 cells/well) were seeded in 6-well plates and transfected with scrambles, miR-335 mimics and anti-miR-335 using Lipofectamine® 2000 Reagent (Invitrogen), respectively.
RNA Transfection
Specific shRNAs that target circ_103973 sh-circ_103973-1 (sh1), and sh-cicr_103973-2 (sh2), negative control (NC), miR-335 inhibitor (anti-miR-335), scramble, siRNAs of PPP6C (si-PPP6C), and si-NC were all designed and provided by GenePharma (Shanghai, China). For cell transfection, HeLa and C33A cells were plated into 6-well plates and cultured at 37°C for 24 hrs, and then cells were transfected with corresponding RNAs using Lipofectamine3000 (Invitrogen, CA, USA) according to the manufacturer’s instructions.
Vector Construction and Transfection
The PPP6C plasmid was purchased from Hanbio (Shanghai, China), and the circ_103973 plasmid was purchased from Genechem (Shanghai, China). Briefly, the extracted DNA was amplified using Taq PCR Master Mix Kit (Tiangen; Cat. no. 201443). The DNA productions were inserted into the pcDNA3.0 vector (Invitrogen) according to previous studies [34, 35]. HeLa and C33A cells (1×105 cells/well) were seeded in 6-well plates and transfected with the circ_103973 or PPP6C overexpression vectors and empty vector (control) using Lipofectamine® 2000 Reagent (Invitrogen).
RNA Extraction and Quantitative Real-Time PCR (RT-PCR) Assay
Total RNAs were isolated CC tissue samples and cell lines using TRIzol reagent (Takara, Tokyo, Japan). RNA quality was examined by a NanoDrop 2000 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA) according to the protocols of manufacturer. Reverse transcription of circ_103973 and PPP6C were carried out using PrimeScript RT reagent kit (Takara, Tokyo, Japan), while the reverse transcription reaction of miR-335 was conducted with a miRNA First Strand Synthesis Kit (Takara, Tokyo, Japan). The qRT-PCR process was conducted on an ABI 7500 fast real-time PCR system (Applied Biosystems, Darmstadt, Germany) with a SYBR Premix Ex Taq Kit (RR420A, Takara, Tokyo). GAPDH was regarded as the endogenous control for circ_103973 and PPP6C; U6 was used as control for miR-335 normalization. Fold changes in expression of RNAs were recorded through 2–ΔΔCt. All sequences of primers are summarized as shown in Table 1.
 |
Table 1 Primer Sequences Used In This Study |
Flow Cytometer Detection
The treated HeLa and C33A cells were collected through centrifugation (1200 ×g for 5 mins), and the cells were counted using hemocytometer and adjusted cell concentration to 1×106/mL. And then cells were fixed with 70% ethanol, treated with RNase A and stained with propidium iodide (20 mg/mL). Finally, the stained cells were all subjected for flow cytometry analysis (EPICS, XL-4, Beckman, CA, USA) following the instructions of manufacturers.
Cell Proliferation Analysis
Effects of circ_103973, miR-335 and PPP6C on cell proliferation of HeLa and C33A cells were evaluated by MTT and colony formation assays. For MTT assay, HeLa and C33A cells were seeded into 96-well plates at a density of 1000 cells/well and cultured overnight at 37°C. Subsequently, 20 µl MTT solution was added into each well and incubated for 4 hrs. After discarding the supernatant, 150 µl DMSO solution was added into each well and incubated for 15 mins at room temperature. Finally, the absorbance of each well was determined at 490 nm. For colony formation assay, treated HeLa and C33A cells were seeded into 6-well plates at a concentration of 3000 cells/well and maintained in a cell incubator with 5% CO2 and 95% O2 for 2 weeks. Subsequently, colonies were fixed using 4% paraformaldehyde followed by Giemsa staining for 30 mins. The visible colonies were counted manually.
Dual Luciferase Reporter Assay
For the establishment of recombinant luciferase reporter plasmids, the wild type (WT) and mutant (mut) fragments of circ_103973 containing putative miR-1205, miR-224, miR-335, miR-526b or miR-630 binding sites were amplified and inserted into the pGL3 (Promega, USA) vector. HeLa cells (2 × 104) were plated into 24-well plates and cultured overnight at 37°C. Then, miR-1205, miR-224, miR-335, miR-526b or miR-630 and Luc-circ_103973-WT or Luc-circ_103973-Mut were co-transfected into HeLa cells using Lipofectamine 3000 reagent. After 48 hrs of co-transfection, the luciferase activities of HeLa cells were measured by the Dual-Luciferase Reporter Assay System (Promega) following the manufacturer’s instructions. The interaction between miR-335 and PPP6C in HeLa cells was also verified following the protocols above.
RNA Pull-Down Assay
HeLa cells stably transfected with biotinylated circ_103973 (Bio-circ_103973) or control (Bio-control) were harvested and lysed. HeLa cell lysates were incubated with C-1 magnetic beads (Life Technologies) at 4°C for 2 hrs followed by the purification by RNeasy Mini Kit (QIAGEN, Duesseldorf, Germany). Finally, qRT-PCR was used to assess the expression of circ_103973 and miR-335.
Statistical Analysis
Statistical analysis was conducted on SPSS v20 software (IBM Corp., Armonk, NY, USA) using one-way analysis of variance. All error bars represent mean ± SEM. P value less than 0.05 was considered as significant.
Results
Circ_103973 was Identified to Be Upregulated in CC
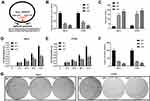
To identify specific circRNAs that might be involved in the tumorigenesis of CC, two circRNA expression profiles (GSE102686 and GSE113696) were downloaded from the GEO database (www.ncbi.nlm.nih.gov/geo). Volcano plots were formed using R software to show the differentially expressed circRNAs in GSE102686 and GSE113696 datasets, and circ_103973 was revealed to be dysregulated in both GSE102686 and GSE113696 datasets (Figure 1A). Results from GEO2R analysis indicated that circ_103973 was highly expressed in CC tissue samples compared to normal samples in GSE102686 and GSE113696 datasets (Figure 1B). To confirm the upregulation of circ_103973, we then detected its expression in both CC clinical tissue samples and cell lines using qRT-PCR. Results indicated that circ_103973 was significantly increased in CC clinical tissues compared to adjacent normal tissues (Figure 1C). Moreover, compared to the human cervical epithelial cell line (HCerEpiC), circ_103973 was also significantly upregulated in four CC cell lines (HeLa, CaSki, C33A, SiHa) (Figure 1D). In addition, we found that CC patients with high circ-103973 expression exhibited a worse prognosis than those patients with low circ_103973 expression (Figure 1E).
Knockdown of circ_103973 Promoted CC Cell Apoptosis and Inhibited CC Cell Proliferation in vitro
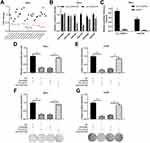
To investigate the biological functions of circ_103973 in CC, we silenced its expression in HeLa and C33A cells by specific shRNAs followed by the analysis of cell apoptosis and proliferation using flow cytometry, MTT and colony formation assays. Two specific shRNAs (sh1 and sh2) were designed to silence the expression of circ_103973 (Figure 2A). qRT-PCR showed a very high knockdown efficiency of circ_103973 shRNAs in HeLa and C33A cells (Figure 2B). For the apoptosis analysis using flow cytometry, circ_103973 shRNAs (sh1 and sh2) treated HeLa and C33A cells showed a significant upregulation of apoptosis rate than those cells treated with NC (Figure 2C). Results from MTT assay showed that circ_103973 shRNAs' treatment resulted in a remarkable inhibition of cell viability of HeLa and C33A cells (Figure 2D and E). In the colony formation assay, we found that circ_103973 knockdown in HeLa and C33A cells could significantly reduce the number of colonies (Figure 2F and G). Moreover, in order to further explore the possible mechanism of circ_103973 knockdown affecting CC cell proliferation, flow cytometer was carried out to the impact of circ_103973 knockdown on cell cycle distribution of HeLa and C33A cells. As shown in Figure S1, the cell percentages in G1 and G2 phases were significantly increased, while the cell percentage in S phase was significantly reduced in circ_103973 siRNAs' transfection group compared to NC group, indicating that knockdown of circ_103973 promoted CC cell cycle arrest.
Circ_103973 Targeted miR-335, Which Reversed the Inhibitory Effects of circ_103973 Knockdown on CC Cell Proliferation
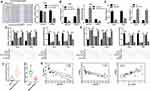
To investigate the molecular mechanism underlying circ_103973 in CC, bioinformatics analysis was performed to screen the target miRNAs of circ_103973 followed by the detection of target miRNAs using qRT-PCR in circ_103973-overexpressed HeLa cells. Results indicated that the change of expression of five miRNAs (miR-1205, miR-224, miR-335, miR-526b and miR-630) was greater than 1-fold in circ_103973-overexpressed HeLa cells compared with empty plasmid-transfected HeLa cells (Figure 3A). In addition, we found that miR-335 showed a lowest level in circ_103973-overexpressed HeLa relative to miR-1205, miR-224, miR-526b and miR-630 levels. Therefore, mir-335 was selected as our research object. Next, dual-luciferase reporter assay showed that miR-224, miR-335, miR-526b and miR-630, but not miR-1205, could attenuate the luciferase activity of HeLa cells driven by Luc-circ_103973-WT (Figure 3B). The luciferase activity of HeLa cells driven by Luc-circ_103973 was not affected by miR-1205, miR-224, miR-335, miR-526b and miR-630. RNA pull-down assay was performed in HeLa cells to further confirm the interaction between circ_103973 and miR-335. Results indicated that the expression of miR-335 in circ_103973 group was significantly higher than that of control group (Figure 3C). Results from MTT assay showed that circ_103973 knockdown significantly inhibited cell viability of HeLa and C33A cells; however, this effect was abrogated by co-transfection of anti-miR-335 and circ_103973 shRNAs (Figure 3D and E). Similarly, the reduced colony number of HeLa and C33A cells induced by circ_103973 was also reversed by co-transfection of anti-miR-335 and circ_103973 shRNAs (Figure 3F and G).
PPP6C was a Target Gene of miR-335 in CC Cell
According to the miRBase prediction, miR-335 could target PPP6C 3ʹ-UTR with a high score; dual-luciferase reporter assay was subsequently used to confirm the relationship between miR-335 and PPP6C in CC cells. The intensity of luciferase in CC cells driven by PPP6C-WT was attenuated by the transfection of miR-335, but not miR-NC; while the luciferase activity in those cells driven by PPP6C-MUT was not influenced by miR-335 or miR-NC (Figure 4A). After transfection of anti-miR-335, the expression of miR-335 was significantly downregulated, while the expression of PPP6C was remarkably upregulated in HeLa cells; after transfection of miR-335 mimics, miR-335 expression was significantly increased, while PPP6C expression was markedly decreased in HeLa cells (Figure 4B). To investigate the roles of PPP6C in circ_103973- and miR-335-mediated effects on TC, we silenced PPP6C by siRNAs and overexpressed PPP6C by overexpression vector, and the knockdown and overexpression efficiencies of si-PPP6C and PPP6C overexpression vector were examined via qRT-PCR assay (Figure 4C). The results of MTT assay revealed that miR-335 mimics transfection could significantly abolish the promoting effects of circ_103973 overexpression on HeLa and C33A cell proliferation, while these effects could be then reversed by the co-transfection of PPP6C in HeLa and C33A cells (Figure 4D, left). Anti-miR-335 treatment could significantly abolish the inhibitory effects of circ_103973 shRNAs on cell proliferation, while these effects could be reversed by the co-transfection of si-PPP6C in HeLa and C33A cells (Figure 4D, right). Similarly, the results of colony formation assay showed similar results with the results of MTT assay (Figure 4E). Data from qRT-PCR indicated that miR-335 was reduced, while PPP6C was increased in the CC tissue specimens compared to normal one (Figure 4F). A negative correlation was observed in the expression of circ_103973 and miR-335, as well as PPP6C and miR-335, in CC, while a positive correlation was revealed between the expression of PPP6C and circ_103973 in CC (Figure 4G).
Discussion
Implication of HPV infection in the etiology of CC has been widely recognized by public; therefore, the invention of HPV vaccine made a remarkable contribution to the primary prevention of CC in the world; however, there are still big challenges in the clinical treatment of CC.14,15 Illuminating the pathogenesis of CC is urgent to explore several effective biomarkers of CC for the screen and treatment of CC patients.
CircRNA was first reported by Sanger HL et al as an abnormal RNA produced by transcription disorder in the 1970s in RNA viruses.16 It did not arouse much attention from the scientific community. However, with the tremendous advancement of high-throughput sequencing and bioinformatics methods, circRNAs have revealed to be expressed extensively in mammalian cells.17 In recent years, circRNAs are being proven to be a critical regulator of multiple biological processes in mammal cells, such as proliferation, apoptosis and differentiation.18 Therefore, circRNAs were considered to be important endogenous biomarkers of human tumors.18 Indeed, an amount of evidences have shown that the expression profiles of circRNAs were dysregulated in various human cancers, such as lung cancer, liver cancer and breast cancer.19 Recently, the biological functions of circRNAs in CC have also been well studied. For instance, circ_8924 was reported by Li W to promote the CC cell proliferation, migration and invasion by sponging miR-518e-5p family and regulating the expression of CBX8.20 Moreover, results from Xianchao Kong el al showed that circ101996 might act as an oncogene of CC through increasing the expression of TPX2 through miR-8075.21 In this study, a circRNA microarray analysis was conducted on two CC-associated circRNA GSE datasets (GSE102686 and GSE113696) downloaded from the GEO database (www.ncbi.nlm.nih.gov/geo) to screen specific circRNAs that might involve in the pathogenesis of CC. Among the different expressed circRNAs, circ_103973 was identified to be significantly increased. After verifying the upregulation of circ_103973 in CC tissues and cell lines, we silenced its expression through specific shRNAs to examine the effects of circ_103973 knockdown on CC cell apoptosis and proliferation in vitro. As results indicated, circ_103973 might act as an oncogene in CC.
According to the hypothesis of competing endogenous RNAs (ceRNA), lncRNAs and circRNAs could sponge miRNAs complying the principle of complementary sequence, resulting in the liberating of gene transcripts that trapped by miRNAs.22,23 Therefore, we conducted bioinformatics analysis to predict the target miRNAs of circ_103973 in the present study, and multiple miRNAs were identified. Of these candidates, miR-335 was selected for further study. MiR-335 was previously reported to be associated with the progression of multiple human tumors, such as lung cancer, colorectal cancer and gastric cancer.24–26 However, up to now, only one paper detected the role of miR-335 family in the pathogenesis of CC, which suggested that lncRNA DANCR could promote CC progression by increasing the expression of ROCK1 through sponging miR-335-5p.27 Similarly, we found that circ_103973 acted as a sponge of miR-335, and inhibition of miR-335 in CC cells could abolish the effects of circ_103973 knockdown on CC cell proliferation. Furthermore, PPP6C was identified as targeted gene of miR-335. PPP6C was reported to be associated with multiple human diseases;28,29 however, its implication in CC remains undetermined. In our study, PPP6C was found to be upregulated in CC, and si-PPP6C could abolish the effects of miR-335 knockdown on circ_103973 silenced CC cells.
Conclusion
In summary, our study demonstrated that increased circ_103973 promoted CC cell proliferation and inhibited CC cell apoptosis by directly increasing the expression of PPP6C through sponging miR-335. The circ_103973/miR-335/PPP6C axis provided a novel insight into the complex molecular mechanisms of CC and revealed that circ_103973 might be a potential diagnostic and therapeutic target of CC. However, additional experiments need to be carried out further to verify the influencing mechanism of circ_103973/miR-335/PPP6C axis on CC cell proliferation, such as cell cycle detection, 3D spheroids analysis and examination of markers related to cell proliferation and cycle. In addition, many in vivo experiments confirm the roles and underlining mechanisms of circ_103973/mir-335/PPP6C axis on CC cell progress including CC xenograft formation.
Ethical Approval and Patient Consent
This study was approved by the ethics committee of the First Affiliated Hospital of Zhejiang University of Traditional Chinese Medicine. All patients signed the written informed consent for this study.
Author Contributions
Conception and intellectual input: ZYP; designed and performed the experiments: ZYP, JXL, ZS, ZQ, JJ and WLC; manuscript drafting: ZYP. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Eiben GL, da Silva DM, Fausch SC, Le Poole IC, Nishimura MI, Kast WM. Cervical cancer vaccines: recent advances in HPV research. Viral Immunol. 2003;16(2):111–121. doi:10.1089/088282403322017866
2. Marth C, Landoni F, Mahner S, et al. Cervical cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2017;28(suppl_4):iv72–iv83. doi:10.1093/annonc/mdx220
3. DeSantis CE, Miller KD, Goding Sauer A, Jemal A, Siegel RL. Cancer statistics for African Americans, 2019. CA Cancer J Clin. 2019;69:211–233. doi:10.3322/caac.v69.3
4. Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189(1):12–19. doi:10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F
5. Yee GP, de Souza P, Khachigian LM. Current and potential treatments for cervical cancer. Curr Cancer Drug Targets. 2013;13(2):205–220. doi:10.2174/1568009611313020009
6. Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids. 2019;16:118–129. doi:10.1016/j.omtn.2019.02.005
7. Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20(12):1829–1842. doi:10.1261/rna.047126.114
8. Liu L, Wang J, Khanabdali R, Kalionis B, Tai X, Xia S. Circular RNAs: isolation, characterization and their potential role in diseases. RNA Biol. 2017;14(12):1715–1721. doi:10.1080/15476286.2017.1367886
9. Holdt LM, Kohlmaier A, Teupser D. Molecular roles and function of circular RNAs in eukaryotic cells. Cell Mol Life Sci. 2018;75(6):1071–1098. doi:10.1007/s00018-017-2688-5
10. He J, Xie Q, Xu H, Li J, Li Y. Circular RNAs and cancer. Cancer Lett. 2017;396:138–144. doi:10.1016/j.canlet.2017.03.027
11. Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14(5):514–521. doi:10.1080/15476286.2015.1122162
12. Wang H, Zhao Y, Chen M, Cui J. Identification of novel long non-coding and circular RNAs in human papillomavirus-mediated cervical cancer. Front Microbiol. 2017;8:1720. doi:10.3389/fmicb.2017.01720
13. Ma HB, Yao YN, Yu JJ, Chen XX, Li HF. Extensive profiling of circular RNAs and the potential regulatory role of circRNA-000284 in cell proliferation and invasion of cervical cancer via sponging miR-506. Am J Transl Res. 2018;10(2):592–604.
14. Chelimo C, Wouldes TA, Cameron LD, Elwood JM. Risk factors for and prevention of human papillomaviruses (HPV), genital warts and cervical cancer. J Infect. 2013;66(3):207–217. doi:10.1016/j.jinf.2012.10.024
15. Wentzensen N, Arbyn M. HPV-based cervical cancer screening –c facts, fiction, and misperceptions. Prev Med. 2017;98:33–35. doi:10.1016/j.ypmed.2016.12.040
16. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73(11):3852–3853. doi:10.1073/pnas.73.11.3852
17. Qu S, Yang X, Li X, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi:10.1016/j.canlet.2015.06.003
18. Zhang Y, Liang W, Zhang P, et al. Circular RNAs: emerging cancer biomarkers and targets. J Exp Clin Cancer Res. 2017;36(1):152. doi:10.1186/s13046-017-0624-z
19. Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev. 2018;48:121–127. doi:10.1016/j.gde.2017.11.007
20. Liu J, Wang D, Long Z, Liu J, Li W. CircRNA8924 promotes cervical cancer cell proliferation, migration and invasion by competitively binding to miR-518d-5p/519-5p family and modulating the expression of CBX8. Cell Physiol Biochem. 2018;48(1):173–184. doi:10.1159/000491716
21. Song T, Xu A, Zhang Z, et al. CircRNA hsa_circRNA_101996 increases cervical cancer proliferation and invasion through activating TPX2 expression by restraining miR-8075. J Cell Physiol. 2019. doi:10.1002/jcp.28128
22. Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–1038. doi:10.1038/nature09144
23. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
24. Yang B, Huang J, Liu H, Guo W, Li G. miR-335 directly, while miR-34a indirectly modulate survivin expression and regulate growth, apoptosis, and invasion of gastric cancer cells. Tumour Biol. 2016;37(2):1771–1779. doi:10.1007/s13277-015-3951-8
25. Liu J, Bian T, Feng J, et al. miR-335 inhibited cell proliferation of lung cancer cells by target Tra2beta. Cancer Sci. 2018;109(2):289–296. doi:10.1111/cas.13452
26. Lu Y, Yang H, Yuan L, et al. Overexpression of miR-335 confers cell proliferation and tumour growth to colorectal carcinoma cells. Mol Cell Biochem. 2016;412(1–2):235–245. doi:10.1007/s11010-015-2630-9
27. LiangH, Zhang C, Guan H, Liu J, Cui Y. LncRNA DANCR promotes cervical cancer progression by upregulating ROCK1 via sponging miR-335-5p. J Cell Physiol. 2019;234(5):7266–7278. doi:10.1002/jcp.27484
28. Shima H, Watanabe T. [Ppp6c deficiency predisposes mouse skin tissue to carcinogenesis]. Seikagaku. 2015;87(5):510–516.
29. Li X, Cai W, Xi W, et al. MicroRNA-31 regulates immunosuppression in ang II (Angiotensin II)-induced hypertension by targeting PPP6C (Protein Phosphatase 6c). Hypertension. 2019;73(5):e14–e24. doi:10.1161/HYPERTENSIONAHA.118.12319
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.