Back to Journals » Cancer Management and Research » Volume 13
Hsa_circ_0043278 Inhibits Tumorigenesis and is Downregulated in Colorectal Cancer
Authors Wang J , Wang T, Hu S, Li J, Ni C, Ye M
Received 3 November 2020
Accepted for publication 14 January 2021
Published 3 February 2021 Volume 2021:13 Pages 965—975
DOI https://doi.org/10.2147/CMAR.S289775
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Jiali Wang,1,2 Tiangong Wang,2 Shiyun Hu,2 Jinyun Li,1 Chao Ni,1 Meng Ye1
1Department of Oncology and Hematology, The Affiliated Hospital of Medical School, Ningbo University, Ningbo, People’s Republic of China; 2School of Medicine, Ningbo University, Ningbo, People’s Republic of China
Correspondence: Meng Ye
Department of Oncology and Hematology, The Affiliated Hospital of Medical School, Ningbo University, 247 Renming Road, Ningbo, Zhejiang, 315000, People’s Republic of China
Tel +86 19857840350
Email [email protected]
Purpose: Circular RNAs are novel endogenous RNAs, which are considered to play a role in tumorigenesis. Nevertheless, the role as well as clinical diagnostic value of most circular RNAs in colorectal cancer are still unclear.
Materials and Methods: We investigated the circular RNA microarray containing expression profiles in samples of colorectal cancer patients by bioinformatics. The consequence indicated that hsa_circ_0043278 was strongly downregulated. We then measured the expression level of hsa_circ_0043278 in tissue samples of colorectal cancer by quantitative real-time polymerase chain reaction. Besides, we also explored the expression condition of the circular RNA in colorectal cancer cell lines including HCT116, SW620, and SW480. Cell counting kit-8, colony formation, and transwell assays, as well as flow cytometry, were applied to detect changes in cell proliferation, migration, apoptosis, and cell cycle progression.
Results: We discovered that circular RNA hsa_circ_0043278 was significantly downregulated in tumor samples (P < 0.0001) as well as cell lines (P < 0.05). The value of the area under the receiver operating characteristic curve was 0.71, with a sensitivity of 0.72 and specificity of 0.70 (P = 0.0006). Moreover, we found that overexpression of hsa_circ_0043278 suppressed proliferation and migratory abilities while promoting apoptosis in colorectal cancer cells.
Conclusion: Our findings revealed that hsa_circ_0043278 inhibited the tumorigenesis of colorectal cancer and could be a potential biomarker for colorectal cancer diagnosis. Besides, it hopes to become a target for treatment.
Keywords: circular RNAs, apoptosis, suppressor, molecular marker, tumorigenesis
Introduction
Colorectal cancer (CRC) is a common type of malignant tumor of the digestive tract. It is the third most common cancer worldwide with 1.48 million people worldwide suffering from CRC, accounting for 9% of new cases in men and 8% in women.1 The global mortality rate of CRC ranks second among malignant tumors.2 The treatment of CRC is mainly surgery combined with radiotherapy, chemotherapy, targeted therapy, and/or immunotherapy. The 5-year and 10-year relative survival rates of CRC patients are 65% and 58%, respectively. If cancer in situ is diagnosed, the 5-year survival rate can reach 90%. However, when the tumor cells spread and metastasize, the 5-year survival rate drops to 11.7%.3 Hence, early diagnosis and treatment are essential.
Circular RNAs (circRNAs) are ubiquitous endogenous RNAs with a closed-loop structure composed of about 30–50 phosphodiester bonds.4 CircRNAs were first discovered in plant viruses by Sanger et al in 1976 and were regarded as by-products of incorrect splicing.5,6 Recently, circRNAs are found to be ubiquitous in various organisms. Salzman et al found that approximately 5.8–23% of human transcribed genes produced circRNAs.7 Compared with linear mRNAs, circRNAs are more resistant to the hydrolysis by exonucleases due to their special covalent loop structure.8 It has been shown that circRNAs participate in many important gene expression regulation processes. They affect the occurrence and development of multiple diseases by acting as sponges of microRNAs (miRNAs) and proteins, transcriptional regulators, and translation templates.9,10 Moreover, the relationship between circRNAs and tumors has received increasing attention, and circRNAs are now considered to be important biomarkers for tumor diagnosis and potential therapeutic targets.11 Recent studies have shown that circRNAs participate in the occurrence and development of CRC by influencing the proliferation, metastasis, and invasive capabilities of tumor cells.12–14
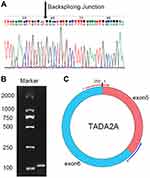
Here, we performed bioinformatic analysis of the dataset GSE126094 acquired from the Gene Expression Omnibus (GEO) database and focused on the 250-nucleotide-long circRNA hsa_circ_0043278, which is located at chr17:35797838–35800763 in the TADA2A gene.15 The circRNA has 39 isoforms formed from backsplicing of TADA2A gene exons, with an alternative ID of hsa_circTADA2A_011. We explored the expression condition of the circRNA within 43 paired samples and its diagnostic potential for CRC. Furthermore, we explored the function of the circRNA in HCT116 and SW620 cell lines.
Materials and Methods
Patients and Samples
Totally, 43 paired CRC tissue samples were acquired from the Gastrointestinal Surgery Department of The Affiliated Hospital of Medical School of Ningbo University (Ningbo, China). All patients received surgery between July 2014 to August 2015 without receiving chemotherapy or radiotherapy. The tissues were stored in RNA preservation solution (Cwbiotech, Beijing, China) at the temperature of −80 °C. The TNM classification of the CRC tissues was identified following the American Joint Committee on Cancer guidelines. All the CRC patients signed the informed consent and agreed to participate in the study. This study was approved by the Human Research Ethical Committee of The Affiliated Hospital of Medical School of Ningbo University. The collection of tissue samples and clinical data of subjects follows the Declaration of Helsinki.
Microarray Assay
To investigate CRC-related circRNAs, dataset GSE126094 that contained circRNA expression levels in 10 CRC patients was downloaded from the GEO database. The expression profiles were normalized and analyzed using R 3.6.2 software. The Bioconductor package “limma” was applied to filter differentially expressed circRNAs.16
Cell Culture
Human kidney 293T cells and human normal colonic epithelial cell line NCM460 as well as CRC cell lines (HCT116, SW620, and SW480) were purchased from the Shanghai Institute of Biochemistry and Cell Biology (Chinese Academy of Sciences, Shanghai, China). 293T, NCM460 and HCT116 cell lines were cultured in Roswell Park Memorial Institute 1640 medium (Corning, Manassas, VA, USA) supplemented with 10% fetal bovine serum (Pan Biotech, Aidenbach, Germany), 50 units/mL penicillin, and 50 μg/mL streptomycin (Gibco, Carlsbad, CA). SW620 and SW480 cell lines were cultured in Leibovitz’s L-15 medium (Gibco) supplemented with 10% fetal bovine serum (Pan Biotech), 50 units/mL penicillin, and 50 μg/mL streptomycin (Gibco). The cell lines were incubated at 37 °C in the atmosphere of 95% air and 5% CO2.
Total RNA Extraction and Reverse Transcription
Total RNA from CRC tissues and cell lines was extracted by TRIzol reagent (Invitrogen, Carlsbad, CA, USA) with the instruction manual. The extracted RNA was then converted into cDNA using the GoScript Reverse Transcription System (Promega Corporation, Fitchburg, WI, USA).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Assay
To detect the relative expression level of circRNA hsa_circ_0043278, qRT-PCR was performed using GoTaq qPCR Master Mix (Promega) by applying a Stratagene MX3005P qPCR System (Agilent Technologies, Santa Clara, CA, USA). Glyceraldehyde-3-phosphate dehydrogenase mRNA was used as internal control. The forward primer for detecting hsa_circ_0043278 for qRT-PCR was 5′-AGCCATTCCATTTCACTACTTCA-3′, with 5′-TCCTGCCAATTTCCAAAGCC-3′ as the reverse primer. ΔCq values were calculated to reflect the expression of hsa_circ_0043278.
Cell Transfection
The hsa_circ_0043278 overexpression plasmid and GV486 vector (GeneChem, Shanghai, China) were used to upregulate the circRNA. The small interfering RNAs (siRNAs) and the corresponding negative control for hsa_circ_0043278 (GenePharma, Shanghai, China) were designed and synthesized to knock down the expression. Lipofectamine 2000 (Invitrogen) along with Opti-MEM I Reduced Serum Medium (Invitrogen) were applied to transfect the plasmid and siRNAs into the cells according to the instruction manual.
Colony Formation Assay
Transfected cells were seeded in 6-well plates at a density of 1.0 × 103 cells per well and incubated for 14 days. After that, each well was washed with phosphate-buffered saline (PBS). Next, 1 mL of 4% paraformaldehyde was added into each well for cell fixation. After 30 min of incubation, paraformaldehyde was removed and 1 mL of 1% crystal violet solution in 10% ethanol was added to stain the colonies. Following staining for 30 min, the colonies were washed with PBS and counted manually.
Cell Counting Kit-8 (CCK-8) Assay
Cell Counting Kit-8 (Dojindo, Tokyo, Japan) was applied to determine cell proliferation levels. Transfected cells at a density of 5.0 × 103 cells per well were seeded in a 96-well plate and then incubated for 24, 48, 72, or 96 hours. CCK-8 reagent was applied according to the instruction manual. The proliferation levels were determined by measuring the absorbance at 450 nm using a SpectraMax M5 Microplate Reader (Molecular Devices, Sunnyvale, CA, USA).
Cell Cycle and Apoptosis Assay
Transfected cells were harvested using 0.25% trypsin-EDTA (Gibco). Then, 70% ethanol was used to fix the harvested cells overnight at the temperature of −20 °C. After fixation, pre-cooled PBS was applied to wash the cells twice. After that, 1 mL of the PI/RNase Staining Buffer (Multi Sciences, Hangzhou, China) was added to stain the cells, which were then resuspended. After 30 min of incubation in the dark, the cells were detected by a FACS Calibur flow cytometer (Becton Dickinson Co., Franklin Lakes, NJ, USA) for cell cycle analysis.
To analyze the extent of apoptosis in the transfected cells, an Annexin V-FITC/PI Apoptosis Kit (Multi Sciences) was used following the instruction manual. A FACS Calibur flow cytometer (Becton Dickinson Co.) was also applied to detect the apoptosis cells.
Transwell Assay
Firstly, PBS was used to wash transfected cells, which were then resuspended in serum-free medium. A 750 μL aliquot of the medium with 20% fetal bovine serum was added to the lower chamber. Then, the cells with 200 μL of serum-free medium was added to the upper chamber at a density of 4×104 per well. Next, after incubation at 37 °C in the atmosphere of 95% air and 5% CO2 for 48 h, the cells passed through the membrane of the upper chamber, which could be stained with crystal violet and observed using a CKCG3 inverted microscope (Olympus, Tokyo, Japan).
Dual-Luciferase Reporter Assay
The dual-luciferase reporter plasmid with hsa_circ_0043278 wild-type and mutant sequence (GenePharma) were co-transfected with miRNA mimics and the corresponding negative control (GenePharma) respectively into 293T cells. The dual-luciferase reporter assay system (Promega) and a SpectraMax M5 Microplate Reader (Molecular Devices) were applied to detect the relative luciferase activity.
Statistical Analysis
SPSS 20.0 software (SPSS Inc., Chicago, IL, USA) was applied to perform statistical analysis. The significance of differences between paired groups was evaluated by the Student’s t-test. The correlation between clinicopathological factors and hsa_circ_0043278 expression level was evaluated by using the chi-squared test. To estimate the diagnostic value of hsa_circ_0043278, the receiver operating characteristic (ROC) curve was analyzed and the area under the curve (AUC) value was calculated. The graphs were plotted by using GraphPad Prism 8 (GraphPad Software Inc., La Jolla, CA, USA).
Results
Microarray Assay
All expression profiles of the 10 CRC samples from the GSE126094 dataset were normalized for further investigation. In total, 291 differentially expressed circRNAs were identified, including 129 downregulated and 162 upregulated circRNAs. Among the differentially expressed circRNAs, hsa_circ_0043278 was particularly strongly downregulated in the 10 CRC samples. Thus, we explored the functions of hsa_circ_0043278 in CRC (Figure 1).
Hsa_circ_0043278 is Downregulated in CRC Tissues and Cell Lines
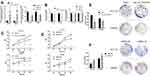
To explore the expression level of hsa_circ_0043278 in CRC tissues, tissue samples from 43 CRC patients were involved. The product of qRT-PCR was analyzed by Sanger sequencing and was found to be consistent with the sequence from circBase (Figure 2).17 Consistent with the result of microarray assay, hsa_circ_0043278 was expressed at a significantly lower level in CRC tissues compared with that in corresponding adjacent normal tissues (P < 0.0001) (Figure 3A). Then, we performed qRT-PCR in three CRC cell lines (HCT116, SW620, and SW480) and in the normal colon epithelial cell line NCM460. As in the case with CRC tissues, hsa_circ_0043278 was also downregulated in CRC cell lines (P = 0.0088, 0.0036, and 0.0463 for HCT116, SW620, and SW480 lines, respectively) (Figure 3B).
Correlation Between Hsa_circ_0043278 and Clinicopathological Factors in CRC Patients
To investigate the association between hsa_circ_0043278 and clinicopathological factors in CRC patients, data about sex, age, tumor differentiation stage, tumor location, tumor size, invasiveness, lymph node metastases, distant metastases, clinical stage, as well as carcinoembryonic antigen and carbohydrate antigen 19-9 levels of CRC patients were retrieved and analyzed. The results suggested that the expression level of hsa_circ_0043278 in CRC tissues was related to tumor size (P = 0.0329), which provided the direction for subsequent cell experiments (Table 1).
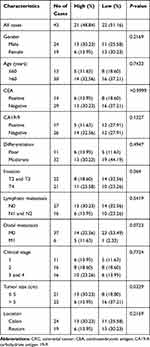 |
Table 1 The Association Between Hsa_circ_0043278 and Clinicopathological Factors in CRC Patients |
Diagnostic Value of Hsa_circ_0043278
We plotted the ROC curve of hsa_circ_0043278. The specificity and sensitivity were calculated to assess the circRNA as a diagnostic indicator. We found that the AUC was 0.71 (P = 0.0006) with the sensitivity of 0.72 and the specificity of 0.70 (Figure 3C). Therefore, hsa_circ_0043278 might have a diagnostic value in CRC.
Hsa_circ_0043278 Inhibits CRC Cell Proliferation
We transfected the hsa_circ_0043278 overexpression plasmid into HCT116 and SW620 cells. The blank vector GV486 was also transfected as negative control (Figure 4A Left). The CCK-8 and colony formation assays showed that hsa_circ_0043278 overexpression significantly inhibited proliferation (Figure 4C) and reduced the numbers of CRC cell colonies compared to those in the negative control groups (Figure 4E). Meanwhile, when hsa_circ_0043278 siRNA was transfected into CRC cells (Figure 4A Right), the resulting hsa_circ_0043278 inhibition promoted proliferation (Figure 4D) and increased the number of CRC cell colonies (Figure 4F) compared with those in the respective negative control groups. The relative expression level of TADA2A in transfected cells was not statistically significant compared with those in the respective negative control groups (Figure 4B).
Hsa_circ_0043278 Promotes CRC Cell Apoptosis
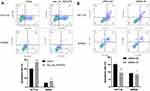
To investigate the influences of hsa_circ_0043278 on the apoptosis in CRC, flow cytometry was performed. We found that hsa_circ_0043278 overexpression promoted apoptosis of both HCT116 and SW620 cells (Figure 5A). Meanwhile, the number of apoptotic cells was reduced by hsa_circ_0043278 siRNA transfection (Figure 5B). Thus, we hypothesize that hsa_circ_0043278 could promote CRC cell apoptosis.
Hsa_circ_0043278 Affects CRC Cell Cycle
Further, we explored whether hsa_circ_0043278 could affect CRC cell cycle. The cell cycle assay showed that hsa_circ_0043278 overexpression blocked HCT116 and SW620 cells in the G0/G1 phase (Figure 6A), whereas CRC cells were arrested in the S phase by hsa_circ_0043278 inhibition (Figure 6B).
Hsa_circ_0043278 Inhibits CRC Cell Migration
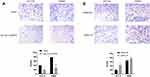
Transwell assay showed that the number of hsa_circ_0043278 overexpressing HCT116 and SW620 cells on the membrane of the chamber was decreased than that in the negative control group (Figure 7A). More cells were observed on the membrane in the groups transfected with hsa_circ_0043278 siRNA than in the respective negative control groups (Figure 7B). Thus, these results indicated that hsa_circ_0043278 inhibited the migration ability of CRC cells.
Hsa_circ_0043278 Sponges with miR-192-3p
Bioinformatics was applied to predict the possible miRNAs that could be sponged by hsa_circ_0043278. According to the result of prediction, a Venn diagram was plotted. From the Venn diagram, we supposed that miR-192-3p might be the most possible miRNA sponged by hsa_circ_0043278 (Figure 8A). To investigate whether hsa_circ_0043278 could sponge with miR-192-3p, we performed dual-luciferase reporter assay in 293T cells. The relative luciferase activity of 293T cells was significantly decreased in cells transfected with hsa_circ_0043278 wildtype sequence and miR-192-3p mimics than mutant sequence (Figure 8B).
Discussion
Thanks to the next-generation sequencing, circRNAs have been found to be involved in a variety of cellular processes. Although they are usually considered as non-coding RNAs, circRNAs are reported to be capable of serving as translation templates.18 A number of studies have found that circRNAs are closely implicated in many diseases, especially cancer.19 For instance, Zhang et al discovered that hsa_circRNA_102002 promoted the epithelial–mesenchymal transition and cell migration in papillary thyroid cancer via the miR-488-3p/HAS2 axis.20 Chen et al found that CircZFR promoted breast cancer progression through the miR-578/HIF1A axis.21 Hong et al demonstrated that circ-CPA4 facilitated tumorigenesis through the miRNA let-7/PD-L1 axis in non-small cell lung cancer.22
Several circRNAs have been associated with CRC progression. Jing et al discovered that hsa_circ_0044556 was upregulated in CRC tissues, where it stimulated the proliferation, migration, and invasiveness of CRC cells.23 Wang et al discovered that hsa_circ_0128846 promoted tumorigenesis by inactivating Hippo/YAP signaling in CRC.24 Chaudhary et al demonstrated that circ-MDM2, which was from the MDM2 locus and identified as DNA damage-regulated circRNA, could inhabit the expression level of p53 and regulate the cell cycle to promote tumorigenesis in CRC.25 Here, we identified hsa_circ_0043278 as a suppressor in CRC, with a significantly low expression level in tissues samples and cell lines. In addition, we demonstrated that hsa_circ_0043278 inhibited the proliferation and migration of HCT116 and SW620 cells while promoting apoptosis. These findings suggest that hsa_circ_0043278 could serve as a potential biomarker for CRC prognosis and be a promising treatment target.
CircRNAs are involved in miRNA sponging that derepresses the translation of miRNA target genes. We hypothesized that hsa_circ_0043278 might also sponge miRNAs to inhibit CRC progression. Thus, we used bioinformatics software to predict the possible miRNAs that could be sponged by hsa_circ_0043278. The most likely candidate, miR-192-3p, has been reported to be associated with CRC, especially with TP53 and KRAS mutations and microsatellite stability. Besides, studies found that miR-192-3p was significantly associated with survival in CRC patients.12 Therefore, we hypothesize that hsa_circ_0043278 inhibits tumorigenesis by sponging miR-192-3p in CRC. Furthermore, we performed dual-luciferase reporter assay to prove this hypothesis.
Conclusion
In conclusion, our study indicates that hsa_circ_0043278 inhibits the proliferation and migration of CRC cells while promoting apoptosis by sponging miR-192-3p, but its expression is downregulated in CRC. Furthermore, the result of our study proposes that hsa_circ_0043278 could become a potential molecular marker for CRC diagnosis and a target for novel CRC treatments.
Acknowledgments
This work was supported by the Zhejiang Provincial Natural Science Foundation of China (grant number LY16H160005).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
3. Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–289. doi:10.3322/caac.21349
4. Chaichian S, Shafabakhsh R, Mirhashemi SM, Moazzami B, Asemi Z. Circular RNAs: a novel biomarker for cervical cancer. J Cell Physiol. 2020;235(2):718–724. doi:10.1002/jcp.29009
5. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73(11):3852–3856. doi:10.1073/pnas.73.11.3852
6. Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7(1):155–160. doi:10.1096/fasebj.7.1.7678559
7. Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32(5):309–316. doi:10.1016/j.tig.2016.03.002
8. Greene J, Baird AM, Brady L, et al. Circular RNAs: biogenesis, function and role in human diseases. Front Mol Biosci. 2017;4:38. doi:10.3389/fmolb.2017.00038
9. Wang Y, Liu J, Ma J, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18(1):116. doi:10.1186/s12943-019-1041-z
10. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
11. Lei B, Tian Z, Fan W, Ni B. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci. 2019;16(2):292–301. doi:10.7150/ijms.28047
12. Slattery ML, Herrick JS, Mullany LE, et al. Colorectal tumor molecular phenotype and miRNA: expression profiles and prognosis. Mod Pathol. 2016;29(8):915–927. doi:10.1038/modpathol.2016.73
13. Xiao H, Liu M. Circular RNA hsa_circ_0053277 promotes the development of colorectal cancer by upregulating matrix metallopeptidase 14 via miR-2467-3p sequestration. J Cell Physiol. 2020;235(3):2881–2890. doi:10.1002/jcp.29193
14. Zhu CL, Sha X, Wang Y, et al. Circular RNA hsa_circ_0007142 is upregulated and targets miR-103a-2-5p in colorectal cancer. J Oncol. 2019;2019:9836819. doi:10.1155/2019/9836819
15. Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41(Database issue):D991–5. doi:10.1093/nar/gks1193
16. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi:10.1093/nar/gkv007
17. Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. Rna. 2014;20(11):1666–1670. doi:10.1261/rna.043687.113
18. Li Z, Ruan Y, Zhang H, Shen Y, Li T, Xiao B. Tumor-suppressive circular RNAs: mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019;110(12):3630–3638. doi:10.1111/cas.14211
19. Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18(1):6. doi:10.1186/s12943-018-0934-6
20. Zhang W, Liu T, Li T, Zhao X. Hsa_circRNA_102002 facilitates metastasis of papillary thyroid cancer through regulating miR-488-3p/HAS2 axis. Cancer Gene Ther. 2020. doi:10.1038/s41417-020-00218-z
21. Chen Z, Wang F, Xiong Y, Wang N, Gu Y, Qiu X. CircZFR functions as a sponge of miR-578 to promote breast cancer progression by regulating HIF1A expression. Cancer Cell Int. 2020;20:400. doi:10.1186/s12935-020-01492-5
22. Hong W, Xue M, Jiang J, Zhang Y, Gao X. Circular RNA circ-CPA4/let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC). J Exp Clin Cancer Res. 2020;39(1):149. doi:10.1186/s13046-020-01648-1
23. Jing L, Wu J, Tang X, et al. Identification of circular RNA hsa_circ_0044556 and its effect on the progression of colorectal cancer. Cancer Cell Int. 2020;20:427. doi:10.1186/s12935-020-01523-1
24. Wang X, Chen Y, Liu W, Liu T, Sun D. Hsa_circ_0128846 promotes tumorigenesis of colorectal cancer by sponging hsa-miR-1184 and releasing AJUBA and inactivating Hippo/YAP signalling. J Cell Mol Med. 2020;24(17):9908–9924. doi:10.1111/jcmm.15590
25. Chaudhary R, Muys BR, Grammatikakis I, et al. A circular RNA from the MDM2 locus controls cell cycle progression by suppressing p53 levels. Mol Cell Biol. 2020;40(9):e00473–19. doi:10.1128/MCB.00473-19
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.