Back to Journals » OncoTargets and Therapy » Volume 13
Hsa_circ_0043265 Suppresses Proliferation, Metastasis, EMT and Promotes Apoptosis in Non-Small Cell Lung Cancer Through miR-25-3p/FOXP2 Pathway
Authors Ren T, Liu C, Hou J, Shan F
Received 18 October 2019
Accepted for publication 26 February 2020
Published 7 May 2020 Volume 2020:13 Pages 3867—3880
DOI https://doi.org/10.2147/OTT.S235231
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Tiejun Ren, Chang Liu, Jianfeng Hou, Fengxiao Shan
Department of Oncology, Luoyang Central Hospital Affiliated to Zhengzhou University, Luoyang, People’s Republic of China
Correspondence: Tiejun Ren
Department of Oncology, Luoyang Central Hospital Affiliated to Zhengzhou University, No. 288, Zhongzhou Road, Luoyang, Henan 471000, People’s Republic of China
Tel +86-379-638922158
Email [email protected]
Purpose: Non-small cell lung cancer (NSCLC) is the largest type of lung cancer (LC) with a higher mortality rate. Circular RNAs (circRNAs) have been shown to play an important role in cancer progression. Therefore, this study was to explore the function of hsa_circ_0043265 in NSCLC.
Methods: The expression levels of hsa_circ_0043265, microRNA-25-3p (miR-25-3p) and forkhead box P2 (FOXP2) were determined by quantitative real-time polymerase chain reaction (qRT-PCR). Ribonuclease R (RNase R) and Actinomycin D (ActD) were used to verify the authenticity and stability of hsa_circ_0043265. Cell counting kit-8 (CCK-8), flow cytometry and transwell assays were used to evaluate the abilities of proliferation, apoptosis, migration and invasion of NSCLC cells. Also, Western blot (WB) analysis was performed to assess the levels of apoptosis, epithelial–mesenchymal transition (EMT) and proliferation-related proteins and FOXP2 protein. RNA immunoprecipitation (RIP) and dual-luciferase reporter assays were used to verify the interaction between miR-25-3p and hsa_circ_0043265 or FOXP2. Besides, mice xenograft models were constructed to confirm the effect of hsa_circ_0043265 on NSCLC tumor growth in vivo.
Results: Hsa_circ_0043265 was lowly expressed in NSCLC tissues and cells, and its overexpression inhibited the proliferation, migration, invasion and EMT process, while improved the apoptosis of NSCLC cells. MiR-25-3p could be sponged by hsa_circ_0043265, and its overexpression could invert the suppression effect of overexpressed-hsa_circ_0043265 on NSCLC progression. Moreover, FOXP2 was a target of miR-25-3p, and its silencing also could reverse the inhibition effect of overexpressed-hsa_circ_0043265 on NSCLC progression. In addition, hsa_circ_0043265 overexpression reduced the tumor growth of NSCLC in vivo.
Conclusion: Hsa_circ_0043265 could sponge miR-25-3p to improve FOXP2 expression, thereby inhibiting NSCLC progression. This study showed that hsa_circ_0043265 could be a potential biomarker for early diagnosis of NSCLC.
Keywords: NSCLC, hsa_ciingrc_0043265, miR-25-3p, FOXP2, progression
Introduction
Non-small cell lung cancer (NSCLC) is a common type of lung cancer (LC), taking up about 80%.1,2 Due to the immaturity of early diagnosis techniques, about 70% of patients are diagnosed with LC in the advanced stage, which greatly increases the death rate of LC.3,4 Despite many proposed treatment strategies for NSCLC, the 5-year survival rate for NSCLC is currently about 17%.5 Therefore, it is urgent to discover new diagnostic markers for NSCLC treatment.
Circular RNAs (circRNAs) are a new type of non-coding RNA with closed-loop structure.6 Current studies have found that circRNAs are abnormally expressed in many cancers, which is expected to be biomarkers for cancer to participate in the regulation of cancer progression.7,8 For example, hsa_circ_0023404 could act as an oncogene to promote the proliferation and metastasis of NSCLC,9 while hsa_circ_0008305 functioned as a tumor suppressor to block the EMT process and tumor metastasis of NSCLC.10 Hsa_circ_0043265 is newly discovered circRNA. Chen et al reported that hsa_circ_0043265 is under-expressed in NSCLC tissues compared to adjacent normal tissues screened by circRNA microarray.11 However, its role and molecular mechanism in NSCLC have not been studied.
MicroRNAs (miRNAs) are small non-coding RNA that bind to target genes to play regulatory roles.12 Some circRNAs can be used as competitive endogenous RNAs (ceRNAs) to sponge miRNAs to take part in the regulation of target genes.13,14 For instance, hsa_circ_0053277 could sponge miR-2467-3p to promote the progression of colorectal cancer throtough regulating MMP14 expression.15 Also, circ_0039569 increased cell growth and metastasis in renal cell carcinoma by the miR-34a-5p/CCL22 axis.16 In cancer-related miRNAs, miR-25-3p had increased expression in many cancers and played a carcinogenic role, such as retinoblastoma, glioma and liver cancer.17–19 Studies had shown that miR-25 was upregulated in NSCLC and participated in the proliferation and metastasis of NSCLC.20–22 Therefore, the study of miR-25-3p can help us better understand the mechanisms that affect the progression of NSCLC.
Forkhead box P2 (FOXP2) is originally discovered as a gene that controls the development of speech and language.23 As more research has progressed, it has been found that FOXP2 is also under-expressed in many cancers. Chen et al revealed that FOXP2 was lower expressed in breast cancer and its knockdown promoted migration and invasion of breast cancer.24 Jia et al suggested that FOXP2 was decreased expression in gastric cancer and took part in the progression of gastric cancer regulated by miR-190.25 Li et al determined that FOXP2 was downregulated in NSCLC and could act as a target of miR-671-3p to participate in the regulation of miR-671-3p on the progression of NSCLC.26 Therefore, FOXP2 might act as an important negative regulator of NSCLC progression.
Herein, our research explored the expression of hsa_circ_0043265 in NSCLC and investigated its role in NSCLC. The discovery of miR-25-3p/FOXP2 axis perfected the molecular mechanism of hsa_circ_0043265 and provided a theoretical basis for the research of hsa_circ_0043265 on the early diagnosis of NSCLC.
Materials and Methods
Samples Collection
The tumor tissues (Tumor) and adjacent normal tissues (Normal) of 25 NSCLC patients were collected from Luoyang Central Hospital Affiliated to Zhengzhou University and frozen at -80°C for experimental use. All patients did not receive any treatment and signed informed consents. This study was authorized by the Ethics Committee of Luoyang Central Hospital Affiliated to Zhengzhou University and was carried out in accordance with the principles of the Declaration of Helsinki.
Cell Culture
NSCLC cells (A549, NCI-H23, NCI-H1299 and HCC827) and human normal lung epithelial cells (BEAS-2B) were bought from American Type Culture Collection (ATCC, Manassas, VA, USA). A549 cells were cultured in Kaighn’s Modification of Ham’s F-12 (F-12K) medium (ATCC), NCI-H23, NCI-H1299 and HCC827 cells were cultured in RPMI-1640 (Sigma-Aldrich, St Louis, MO, USA) and BEAS-2B cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; HyClone, Logan, Utah, USA). All medium contained 10% fetal bovine serum (FBS; Sijiqing, Hangzhou, China), 1% penicillin (100 U/mL)/streptomycin (100 μg/mL), and all cells were incubated at 37°C with 5% CO2.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNAs were extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and reverse-transcribed into complementary DNA (cDNA) using Universal RT-PCR Kit (Solarbio, Beijing, China). QRT-PCR was analyzed by the ABI 7900 System (Applied Biosystems, Foster City, CA, USA) using SYBR Green (Solarbio). Relative expression values were calculated by the 2−∆∆Ct method and normalized by glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or U6. All primers were listed as below: hsa_circ_0043265: F 5ʹ-CAACGCAGGCATCAGAAGATT-3ʹ, R 5ʹ-AGGAAGGCCACTTCATAAGTCTG-3ʹ; FOXP2: F 5ʹ-CCACGAAGACCTCAATGGTT-3ʹ, R 5ʹ-TCACGCTGAGGTTTCACAAG-3ʹ; GAPDH: F 5ʹ-AGAAGGCTGGGGCTCATTTG-3ʹ, R 5ʹ-AGGGGCCATCCACAGTCTTC-3ʹ; miR-25-3p: F 5ʹ-ACGTCTACACTCCATTGCACTTGTCTCGG-3ʹ, R 5ʹ-CAGTGCGTGTCGTGGAGT-3ʹ; U6: F 5ʹ-GCAGGAGGTCTTCACAGAGT-3ʹ, R 5ʹ-TCTAGAGGAGAAGCTGGGGT-3ʹ.
Chromogenic in situ Hybridization (CISH)
About 4 μm-thick formalin-fixed paraffin-embedded sections of NSCLC tumor tissues (Tumor, n = 6) and adjacent normal tissues (Normal, n = 6) from the original cohort of 25 NSCLC patients were used for mRNA CISH staining, using the Leica Bond Rx autostainer and detected using the ViewRNA red stain kit (Affymetrix, Santa Clara, CA, USA). Probe for hsa_circ_0043265 was obtained from Affymetrix.
CircRNA Identification
The extracted RNA was treated with Ribonuclease R (RNase R; Duma, Shanghai, China) and the hsa_circ_0043265 expression was detected by qRT-PCR to verify the authenticity of hsa_circ_0043265. GAPDH was served as the endogenous control. Also, A549 and NCI-H23 cells were treated with Actinomycin D (ActD; Amyjet Scientific, Wuhan, China) for 0 h, 5 h, 10 h, 15 h and 20 h. The expression levels of hsa_circ_0043265 and GAPDH were determined by qRT-PCR to verify the stability of hsa_circ_0043265.
Plasmid Construction and Transfection
Hsa_circ_0043265 overexpression plasmid and small interfering RNA (siRNA) (hsa_circ_0043265 and si-hsa_circ_0043265) or their negative controls (circ-NC and si-NC), miR-25-3p mimic (miR-25-3p) and its negative control (NC), siRNA against FOXP2 (si-FOXP2) and its negative control (scramble) were provided by Ribobio (Guangzhou, China). The sequences of siRNA and miRNA mimic were listed as follows: si-hsa_circ_0043265: 5ʹ-AAACAAAGAGAUAGCAUUGCC-3ʹ; si-FOXP2: 5ʹ-GACAGGCAGTTAACACTTAAT-3ʹ; si-NC or scramble: 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ; miR-25-3p mimic: 5ʹ-CAUUGCACUUGUCUCGGUCUGA-3ʹ; NC: 5ʹ-GGUUCGUACGUACACUGUUCA-3ʹ. Lentivirus hsa_circ_0043265 overexpression vector (Lv-hsa_circ_0043265) and its negative control (Lv-NC) were constructed by Genewiz (Suzhou, China). Lipofectamine 2000 (Invitrogen) was used to transfect all plasmid vectors into A549 and NCI-H23 cells.
Cell Counting Kit-8 (CCK-8) Assay
A549 and NCI-H23 cells were seeded into 96-well plates. After transfection, cells were treated with CCK-8 solution (Amyjet Scientific) for 4 h at the specified time point (24 h, 48 h and 72 h). The absorbance was measured at 450 nm using a microplate reader (Molecular devices, Shanghai, China).
Flow Cytometry
A549 and NCI-H23 cells were digested with trypsin (Solarbio) and collected into the centrifuge tubes. After centrifugation, the cells were re-suspended with binding buffer (Beyotime, Shanghai, China) and then stained with Annexin V-fluorescein isothiocyanate (FITC) and Propidium iodide (PI) (Beyotime) for 15 min. Fluorescence signals were detected by Flow cytometer (Beckman Coulter, San Jose, CA, USA) and the apoptosis rate of cells was counted.
Transwell Assay
The migration and invasion of A549 and NCI-H23 cells were detected using transwell chambers with an 8 µm pore size (Corning lnc., Corning, NY, USA), which coated Matrigel (Corning lnc.) to detect cell invasion and not coated Matrigel to detect cell migration. Briefly, A549 and NCI-H23 cells were seeded into upper chambers containing the serum-free medium. The lower chambers were filled with serum medium. After 24 h, the lower chamber cells were fixed with paraformaldehyde and stained with crystal violet, and the numbers of migration and invasion cells were examined under an inverted microscope (Leica, Wetzlar, Germany).
Western Blot (WB) Analysis
Total proteins were extracted using RIPA buffer (Beyotime) and quantified using BCA Kit (Beyotime). The equivalent amount of protein was isolated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Then, the membranes were closed with 5% non-fat milk and incubated with the primary antibodies against caspase 3 (1:1000, Beyotime), B-cell lymphoma-2 (Bcl-2; 1:2000, Abcam, Cambridge, MA, USA), Bcl-2-associated x protein (Bax; 1:5000, Abcam), E-cadherin (1:1000, Abcam), Vimentin (1:2000, Abcam), FOXP2 (1:1000, Abcam), proliferating cell nuclear antigen (PCNA; 1:5000, Abcam) or GAPDH (1:5000, Abcam) at 4°C overnight. After incubated with the secondary antibody (1:2000, Abcam) for 1 h, the membranes were treated with enhanced chemiluminescence reagent (Beyotime) to visualize the protein signals.
RNA Immunoprecipitation (RIP) Assay
Magna RIPTM RNA-Binding Protein Immunoprecipitation Kit (Millipore) was used for RIP assay. A549 and NCI-H23 cells were lysed using RIP lysis buffer and incubated with magnetic beads conjugated with the argonaute2 (Ago2) antibody (Anti-Ago2) or negative control immunoglobulin G (IgG) antibody (Anti-IgG). Part of cell lysates was not incubated with magnetic beads and served as a blank control (Input). After purification of RNA from the immunoprecipitates, the enrichment of hsa_circ_0043265, miR-25-3p and FOXP2 in Input, Anti–IgG or Anti-Ago2 were analyzed by qRT-PCR.
Dual-Luciferase Reporter Assay
ENCORI tool was used to predict the binding sites between miR-25-3p and hsa_circ_0043265 or FOXP2. The wild-type and mutant-type hsa_circ_0043265 (hsa_circ_0043265-wt/mut) or FOXP2 3ʹUTR (FOXP2-wt/mut) reporter vectors containing the miR-25-3p target binding sites and mutant binding sites were synthesized by General Biosystems (Anhui, China). A549 and NCI-H23 cells were co-transfected with miR-25-3p mimic or miR-NC and the above reporter vectors using Lipofectamine 2000 (Invitrogen). After transfection for 48 h, the luciferase activities were measured using Dual-Lucy Assay Kit (Solarbio).
Animal Experiments
Animal experiments were authorized by the Animal Research Committee of Luoyang Central Hospital Affiliated to Zhengzhou University. One-month-old male BALB/c-nude mice were brought from Cavens Laboratory Animals Co., LTD. (Changzhou, China). A549 cells transfected with Lv-hsa_circ_0043265 or Lv-NC were injected into nude mice to establish the mice xenograft models. Tumor length and width were recorded weekly to calculate the tumor volume according to the formula: volume= length × width2/2. After 5 weeks, the tumor was removed for weight measurement, qRT-PCR and WB analysis.
Statistical Analysis
The data were presented as the means ± standard deviation (SD) and analyzed using the SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA). Statistical significance was determined using Student’s t-test and one-way analysis of variance. Pearson correlation coefficient analysis was used to evaluate the correlation. Differences were defined as statistically significant for P < 0.05.
Results
Hsa_circ_0043265 Was Downregulated in NSCLC Tissues and Cells
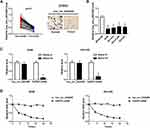
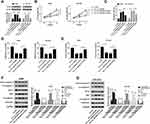
Firstly, we detected the expression of hsa_circ_0043265 in NSCLC. As shown in Figure 1A, hsa_circ_0043265 expression was significantly lower in NSCLC tumor tissues (Tumor) than that in adjacent normal tissues (Normal), and CISH confirmed the downregulated expression of hsa_circ_0043265 in NSCLC tissues. Besides, the relationship between hsa_circ_0043265 expression and clinicopathological parameters of NSCLC patients showed that low hsa_circ_0043265 expression was positively correlated with tumor size, tumor nodes metastasis (TNM) stage and lymph node metastasis in NSCLC patients (P < 0.05, Table 1). Compared with BEAS-2B cells, the expression of hsa_circ_0043265 in four NSCLC cells (A549, NCI-H23, NCI-H1299 and HCC827) was dramatically decreased, especially in A549 and NCI-H23 cells (Figure 1B). To verify the authenticity of hsa_circ_0043265, we treated A549 and NCI-H23 cells with RNase R. The results showed that RNase R treatment markedly reduced the expression of linear transcript GAPDH, while the expression of hsa_circ_0043265 remained unchanged in A549 and NCI-H23 cells (Figure 1C). Also, we used ActD to verify the stability of hsa_circ_0043265 and found that hsa_circ_0043265 was more stable than GAPDH in A549 and NCI-H23 cells (Figure 1D). These results suggested that hsa_circ_0043265 was a real circRNA and might have an essential function in NSCLC.
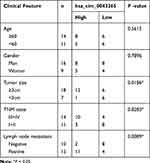 |
Table 1 Correlation Between hsa_circ_0043265 Expression and Clinicopathological Parameters of NSCLC Patients (n = 25) |
Overexpressed-hsa_circ_0043265 Inhibited Proliferation, Migration, Invasion, EMT and Induced Apoptosis in NSCLC Cells
To explore the biological effect of hsa_circ_0043265 on NSCLC, we transfected hsa_circ_0043265 overexpression plasmid into A549 and NCI-H23 cells. QRT-PCR results indicated that hsa_circ_0043265 overexpression plasmid had an excellent promotion effect on hsa_circ_0043265 expression in both cells, indicating a good transfection efficiency (Figure 2A). Then, CCK-8 assay results revealed that compared with circ-NC, overexpression of hsa_circ_0043265 remarkably hindered the proliferation of A549 and NCI-H23 cells (Figure 2B). Besides, through flow cytometry, we also found that overexpressed-hsa_circ_0043265 promoted the apoptosis rates of A549 and NCI-H23 cells, indicating that hsa_circ_0043265 overexpression induced the apoptosis of NSCLC cells (Figure 2C). Furthermore, the number of migrated and invaded A549 and NCI-H23 cells were markedly reduced after overexpression of hsa_circ_0043265, suggesting that hsa_circ_0043265 overexpression suppressed the migration and invasion of NSCLC cells (Figure 2D and E). By detecting the levels of apoptosis and EMT-related proteins, we concluded that hsa_circ_0043265 overexpression increased the protein levels of C-caspase 3/total-caspase 3, Bax, E-cadherin, and reduced Bcl-2 and Vimentin protein levels (Figure 2F), indicating that overexpressed-hsa_circ_0043265 promoted the apoptosis and impeded the EMT process of A549 and NCI-H23 cells. Therefore, the above results confirmed that hsa_circ_0043265 might be involved in the development of NSCLC as a tumor suppressor.
miR-25-3p Had a Target Site with hsa_circ_0043265
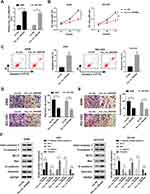
Moreover, we also discovered that miR-25-3p was upregulated in NSCLC tissues and cells (Figure 3A and B), and its expression trend was opposite to that of hsa_circ_0043265. To confirm the relationship between them, RIP assay was carried out and the expression levels of endogenous hsa_circ_0043265 and miR-25-3p pulled down by Ago2 were analyzed by qRT-PCR. The results showed that Anti-Ago2 precipitated a large number of hsa_circ_0043265 and miR-25-3p in A549 and NCI-H23 cells, indicating that hsa_circ_0043265 might be related to miR-25-3p (Figure 3C). Then, through the ENCORI tool, we found that miR-25-3p had complementary binding sites with hsa_circ_0043265 (Figure 3D). Besides, correlation analysis showed that the expression of miR-25-3p was negatively correlated with hsa_circ_0043265 in NSCLC tissues (Figure 3E). To further confirm the interaction between them, we performed the dual-luciferase reporter assay, which showed that miR-25-3p overexpression remarkably reduced the luciferase activity of hsa_circ_0043265-wt in A549 and NCI-H23 cells, whereas had no effect on hsa_circ_0043265-mut (Figure 3F). Furthermore, in order to evaluate the effect of hsa_circ_0043265 expression on miR-25-3p expression in NSCLC cells, we transfected hsa_circ_0043265 overexpression plasmid or si-hsa_circ_0043265 into A549 and NCI-H23 cells to detect the expression of miR-25-3p expression, and discovered that miR-25-3p expression was impeded by hsa_circ_0043265 overexpression, while improved by hsa_circ_0043265 knockdown (Figure 3G). All data demonstrated that hsa_circ_0043265 could sponge miR-25-3p in NSCLC cells.
miR-25-3p Could Reverse the Effects of Overexpressed-hsa_circ_0043265 on Proliferation, Apoptosis, Migration, Invasion and EMT in NSCLC Cells
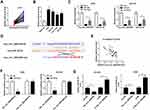
To confirm the function of miR-25-3p on the progression of NSCLC cells, we co-transfected hsa_circ_0043265 overexpression plasmid and miR-25-3p mimic into A549 and NCI-H23 cells. By detecting the expression of miR-25-3p, we found that overexpressed-hsa_circ_0043265 had a good inhibitory effect on miR-25-3p expression and miR-25-3p mimic had a good promoting effect on miR-25-3p expression, indicating that the transfection of them was successful (Figure 4A). Besides, CCK-8 assay and flow cytometry results determined that the inhibitory effect of hsa_circ_0043265 overexpression on proliferation and promotion effect of it on apoptosis in A549 and NCI-H23 cells could be reversed by miR-25-3p overexpression (Figure 4B and C). Also, transwell assay revealed that miR-25-3p overexpression inverted the inhibition effect of overexpressed-hsa_circ_0043265 on the migration and invasion of A549 and NCI-H23 cells (Figure 4D and E). Furthermore, the promotion effects of overexpressed-hsa_circ_0043265 on the protein levels of C-caspase 3/total-caspase 3, Bax, E-cadherin and the suppression effects on the protein levels of Bcl-2 and Vimentin could be restored by miR-25-3p overexpression (Figure 4F and G). Hence, these results suggested that miR-25-3p played a vital role in the regulation of NSCLC progression regulated by hsa_circ_0043265.
miR-25-3p Targeted FOXP2
For another, we detected the FOXP2 expression in NSCLC. QRT-PCR results indicated that FOXP2 was lower expressed in NSCLC tumor tissues (Tumor) compared with adjacent normal tissues (Normal) (Figure 5A). Similarly, at the protein level, we get consistent results (Figure 5B). Besides, we found that FOXP2 protein level was markedly reduced in NSCLC cells compared with BEAS-2B cells (Figure 5C). These revealed that the expression trend of FOXP2 was opposite to that of miR-25-3p, so we further verified the relationship between them. RIP assay showed that miR-25-3p and FOXP2 were enriched in Anti-Ago2, suggesting that FOXP2 might be related to miR-25-3p in NSCLC cells (Figure 5D and E). With the help of ENCORI tool, we found a putative miR-25-3p binding site located in FOXP2 3ʹUTR (Figure 5F). Also, correlation analysis determined that the FOXP2 expression was positively correlated with hsa_circ_0043265 and negatively correlated with miR-25-3p in NSCLC tissues (Figure 5G and H). Dual-luciferase reporter assay results revealed that miR-25-3p overexpression markedly inhibited the luciferase activity of FOXP2-wt, while did not affect FOXP2-mut in A549 and NCI-H23 cells (Figure 5I). To determine the regulatory effect of hsa_circ_0043265 and miR-25-3p on FOXP2, we detected the protein level of FOXP2 in A549 and NCI-H23 cells co-transfected with hsa_circ_0043265 overexpression plasmid and miR-25-3p mimic. The results indicated that miR-25-3p overexpression could restrain the FOXP2 level, while the addition of hsa_circ_0043265 reversed this inhibition and thus restored FOXP2 level (Figure 5J). Therefore, these data revealed that FOXP2 was a target of miR-25-3p, and its expression was regulated by miR-25-3p and hsa_circ_0043265.
Knockdown of FOXP2 Inverted the Effects of Overexpressed-hsa_circ_0043265 on Proliferation, Apoptosis, Migration, Invasion and EMT in NSCLC Cells
To identify the role of FOXP2 in the progression of NSCLC cells, we co-transfected hsa_circ_0043265 overexpression plasmid and si-FOXP2 into A549 and NCI-H23 cells. The detection of FOXP2 protein level results showed that FOXP2 level was promoted by hsa_circ_0043265 overexpression and suppressed by FOXP2 knockdown in A549 and NCI-H23 cells, indicating that the transfection of them was successful (Figure 6A). CCK-8, flow cytometry and transwell assays results determined that the inhibition effects of overexpressed-hsa_circ_0043265 on the proliferation, migration, invasion and the promotion effect of it on the apoptosis of A549 and NCI-H23 cells could be reversed by FOXP2 silencing (Figure 6B–E). Besides, the decrease of C-caspase 3/total-caspase 3 and Bax protein levels and the increase of Bcl-2 level also confirmed that FOXP2 silencing reversed the promoting effect of hsa_circ_0043265 overexpression on apoptosis of A549 and NCI-H23 cells (Figure 6F and G). Then, E-cadherin and Vimentin levels were detected in A549 and NCI-H23 cells, and the results disclosed that the suppression of hsa_circ_0043265 overexpression on the EMT process in A549 and NCI-H23 cells could be inverted by FOXP2 knockdown (Figure 6F and G). These results illustrated that FOXP2 was an essential target gene in the regulation of hsa_circ_0043265 on the progression of NSCLC cells.
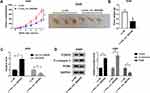
Overexpression of hsa_circ_0043265 Inhibited the Tumor Growth of NSCLC in vivo
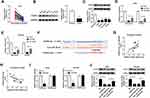
To further explore the influence of hsa_circ_0043265 on NSCLC, we constructed the mice xenograft models. After 5 weeks, we discovered that the tumor volume and weight of the hsa_circ_0043265 overexpression group were markedly reduced compared with the Lv-NC group (Figure 7A and B). Moreover, qRT-PCR showed that hsa_circ_0043265 expression was remarkably increased in the Lv-hsa_circ_0043265 group compared with the Lv-NC group (Figure 7C), indicating that the transfection effect was excellent. Also, we uncovered that miR-25-3p expression was suppressed while the FOXP2 protein level was increased in the Lv-hsa_circ_0043265 group (Figure 7C and D). At the same time, C-caspase 3 was improved while proliferation-related protein PCNA level was inhibited by overexpressed-hsa_circ_0043265 (Figure 7D), suggesting that hsa_circ_0043265 overexpression promoted apoptosis and inhibited proliferation in the tumor. Therefore, the above results confirmed that hsa_circ_0043265 had an anti-cancer effect in NSCLC.
Discussion
CircRNAs currently play a vital role in cancer progression, and many new circRNAs are waiting to be explored. In our study, we confirmed that hsa_circ_0043265 was lowly expressed in NSCLC, which was consistent with the results of Chen’s microarray analysis.11 Also, gain-of-functional experiment results determined that elevated expression of hsa_circ_0043265 impeded proliferation, migration, invasion, EMT, and induced apoptosis in NSCLC. Besides, overexpressed-hsa_circ_0043265 also reduced the tumor volume and weight of NSCLC in vivo.
At present, many miRNAs have been proved to play an oncogene or tumor suppressor role in NSCLC. Ren et al revealed that miR-210-3p was upregulated in NSCLC and promoted proliferation and inhibited apoptosis of NSCLC cells.27 Moreover, Huang et al suggested that miR-497 could act as a tumor suppressor in NSCLC to hinder the proliferation and migration of NSCLC cells.28 For miR-25-3p, Ding et al reported that miR-25 could promote the migration and invasion of NSCLC.20 Also, Xiang et al suggested that miR-25 was related to the proliferation and motility of NSCLC.21 All these confirmed that miR-25 might be a vital regulator of NSCLC and played an active role in the development of NSCLC. Here, we discovered that miR-25-3p expression was increased in NSCLC. After bioinformatics prediction and further experimental verification, we determined that miR-25-3p could bind to hsa_circ_0043265, and its expression was regulated by hsa_circ_0043265. Furthermore, the inhibition effect of hsa_circ_0043265 overexpression on NSCLC progression could be reversed by overexpressed-miR-25-3p. In vivo experiment, we also verify that elevated expression of hsa_circ_0043265 could reduce miR-25-3p expression. Given the role of miR-25-3p expression in promoting the progression of NSCLC, the conclusion that hsa_circ_0043265 promoted the progression of NSCLC by absorbing miR-25-3p could be better understood.
FOXP2 has been reported as a tumor inhibitor, and its downregulation was involved in many cancers progression.24,25 Yu et al reported that miR-196b promoted the migration and invasion of hepatocellular carcinoma through targeting FOXP2.29 Another study indicated that miR-23a functioned as an oncogene to promote pancreatic carcinoma progression by targeting FOXP2.30 In NSCLC, FOXP2 expression was also considered negatively correlated with the progression of NSCLC.26 Consistent with previous studies, we also found that FOXP2 expression was decreased in NSCLC. The function of miRNAs is achieved by regulating the expression of target genes.31,32 Here, RIP and dual-luciferase reporter assays confirmed that FOXP2 was a potential target gene for miR-25-3p. FOXP2 expression was regulated by miR-25-3p and hsa_circ_0043265. Besides, functional experiments confirmed that FOXP2 knockdown also could invert the inhibitory of hsa_circ_0043265 overexpression on the progression of NSCLC cells. In addition, overexpressed-hsa_circ_0043265 improved FOXP2 expression in vivo. All results were consistent with previous reports that FOXP2 still acted as a tumor suppressor in NSCLC, so it act as a target gene for miR-25-3p to involve in the regulation of hsa_circ_0043265 on NSCLC progression might help us better understand the mechanism of hsa_circ_0043265.
Of course, our study has some limitations. The overexpression of miR-25-3p only partially reverses the function of hsa_circ_0043265, which indicates that there are other miRNAs involved in the regulation of hsa_circ_0043265 on NSCLC progression. Similarly, FOXP2 knockdown also partially inverts the effect of hsa_circ_0043265, which indicates that miR-25-3p also has other target genes involved in the regulation of hsa_circ_0043265 on NSCLC progression, which therefore needs further exploration and improvement.
Conclusion
In summary, we concluded that hsa_circ_0043265 regulated FOXP2 expression to hinder the proliferation, metastasis, EMT and accelerate the apoptosis of NSCLC cells through sponging miR-25-3p. The study of hsa_circ_0043265/miR-25-3p/FOXP2 pathway provided new targets for early diagnosis of NSCLC.
Highlights
- Overexpression of hsa_circ_0043265 inhibits NSCLC progression and tumor growth;
- MiR-25-3p can be sponged by hsa_circ_0043265;
- FOXP2 is a target of miR-25-3p.
Disclosure
The authors declare that they have no financial conflicts of interest in this work.
References
1. McIntyre A, Ganti AK. Lung cancer-A global perspective. J Surg Oncol. 2017;115(5):550–554. doi:10.1002/jso.24532
2. Ettinger DS, Akerley W, Borghaei H, et al. Non-small cell lung cancer, version 2.2013. J Natl Compr Canc Netw. 2013;11(6):
3. Molina JR, Adjei AA, Jett JR. Advances in chemotherapy of non-small cell lung cancer. Chest. 2006;130(4):1211–1219. doi:10.1378/chest.130.4.1211
4. Williams CD, Gajra A, Ganti AK, Kelley MJ. Use and impact of adjuvant chemotherapy in patients with resected non-small cell lung cancer. Cancer. 2014;120(13):1939–1947. doi:10.1002/cncr.28679
5. Ma PC. Personalized targeted therapy in advanced non-small cell lung cancer. Cleve Clin J Med. 2012;79 Electronic Suppl 1:eS56–60. doi:doi:10.3949/ccjm.79.s2.12
6. Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi:10.1016/j.canlet.2015.06.003
7. Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi:10.1186/s12943-017-0663-2
8. Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi:10.1016/j.cca.2015.02.018
9. Liu C, Zhang Z, Qi D. Circular RNA hsa_circ_0023404 promotes proliferation, migration and invasion in non-small cell lung cancer by regulating miR-217/ZEB1 axis. Onco Targets Ther. 2019;12:6181–6189. doi:10.2147/OTT.S201834
10. Wang L, Tong X, Zhou Z, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-beta-induced epithelial-mesenchymal transition and metastasis by controlling TIF1gamma in non-small cell lung cancer. Mol Cancer. 2018;17(1):140. doi:10.1186/s12943-018-0889-7
11. Chen T, Yang Z, Liu C, et al. Circ_0078767 suppresses non-small-cell lung cancer by protecting RASSF1A expression via sponging miR-330-3p. Cell Prolif. 2019;52(2):e12548. doi:10.1111/cpr.12548
12. Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. doi:10.1016/j.addr.2015.05.001
13. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
14. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi:10.1038/nature12986
15. Xiao H, Liu M. Circular RNA hsa_circ_0053277 promotes the development of colorectal cancer by upregulating matrix metallopeptidase 14 via miR-2467-3p sequestration. J Cell Physiol. 2019. doi:10.1002/jcp.29193
16. Jin C, Shi L, Li Z, et al. Circ_0039569 promotes renal cell carcinoma growth and metastasis by regulating miR-34a-5p/CCL22. Am J Transl Res. 2019;11(8):4935–4945.
17. Chen H, Pan H, Qian Y, Zhou W, Liu X. MiR-25-3p promotes the proliferation of triple negative breast cancer by targeting BTG2. Mol Cancer. 2018;17(1):4. doi:10.1186/s12943-017-0754-0
18. Wan W, Wan W, Long Y, et al. MiR-25-3p promotes malignant phenotypes of retinoblastoma by regulating PTEN/Akt pathway. Biomed Pharmacother. 2019;118:109–111. doi:10.1016/j.biopha.2019.109111
19. Peng G, Yang C, Liu Y, Shen C. miR-25-3p promotes glioma cell proliferation and migration by targeting FBXW7 and DKK3. Exp Ther Med. 2019;18(1):769–778. doi:10.3892/etm.2019.7583
20. Ding X, Zhong T, Jiang L, Huang J, Xia Y, Hu R. miR-25 enhances cell migration and invasion in non-small-cell lung cancer cells via ERK signaling pathway by inhibiting KLF4. Mol Med Rep. 2018;17(5):7005–7016. doi:10.3892/mmr.2018.8772
21. Xiang J, Hang JB, Che JM, Li HC. MiR-25 is up-regulated in non-small cell lung cancer and promotes cell proliferation and motility by targeting FBXW7. Int J Clin Exp Pathol. 2015;8(8):9147–9153.
22. Liu B, Sun X. miR-25 promotes invasion of human non-small cell lung cancer via CDH1. Bioengineered. 2019;10(1):271–281. doi:10.1080/21655979.2019.1632668
23. Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413(6855):519–523. doi:10.1038/35097076
24. Chen MT, Sun HF, Li LD, et al. Downregulation of FOXP2 promotes breast cancer migration and invasion through TGFbeta/SMAD signaling pathway. Oncol Lett. 2018;15(6):8582–8588. doi:10.3892/ol.2018.8402
25. Jia WZ, Yu T, An Q, et al. MicroRNA-190 regulates FOXP2 genes in human gastric cancer. Onco Targets Ther. 2016;9:3643–3651. doi:10.2147/OTT.S103682
26. Li ZY, Zhang ZZ, Bi H, et al. Upregulated microRNA6713p promotes tumor progression by suppressing forkhead box P2 expression in non-small-cell lung cancer. Mol Med Rep. 2019;20(4):3149–3159. doi:10.3892/mmr.2019.10563
27. Ren J, Li X, Dong H, et al. miR-210-3p regulates the proliferation and apoptosis of non-small cell lung cancer cells by targeting SIN3A. Exp Ther Med. 2019;18(4):2565–2573. doi:10.3892/etm.2019.7867
28. Huang Q, Li H, Dai X, Zhao D, Guan B, Xia W. miR-497 inhibits the proliferation and migration of A549 non-small-cell lung cancer cells by targeting FGFR1. Mol Med Rep. 2019;20(4):3959–3967. doi:10.3892/mmr.2019.10611
29. Yu Z, Lin X, Tian M, Chang W. microRNA196b promotes cell migration and invasion by targeting FOXP2 in hepatocellular carcinoma. Oncol Rep. 2018;39(2):731–738. doi:10.3892/or.2017.6130
30. Diao H, Ye Z, Qin R. miR-23a acts as an oncogene in pancreatic carcinoma by targeting FOXP2. J Investig Med. 2018;66(3):676–683. doi:10.1136/jim-2017-000598
31. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/s0092-8674(04)00045-5
32. Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9(3):219–230. doi:10.1038/nrm2347
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.