Back to Journals » Cancer Management and Research » Volume 13
Hsa_circ_0015326 Promotes the Proliferation, Invasion and Migration of Ovarian Cancer Through miR-127-3p/MYB
Authors Zhang C, Liu W, Li F, Feng Y, Li Y, Wang J
Received 9 November 2020
Accepted for publication 17 February 2021
Published 10 March 2021 Volume 2021:13 Pages 2265—2277
DOI https://doi.org/10.2147/CMAR.S291218
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Beicheng Sun
This paper has been retracted.
Cuiying Zhang, 1 Wei Liu, 2 Fei Li, 1 Yang Feng, 1 Yunyun Li, 1 Jia Wang 3
1Department of Gynaecology, Yongchuan Hospital of Chongqing Medical University, Chongqing, People’s Republic of China; 2Department of Orthopedics, Yongchuan Hospital of Chongqing Medical University, Chongqing, People’s Republic of China; 3Department of Gynaecology and Obstetrics, University-Town Hospital of Chongqing Medical University, Chongqing, People’s Republic of China
Correspondence: Jia Wang
Department of Gynaecology and Obstetrics, University-Town Hospital of Chongqing Medical University, No. 55 University Town Middle Road, Shapingba District, Chongqing, 401331, People’s Republic of China
Tel +86 13508307321
Fax +86 023-65715700
Email [email protected]
Background: More and more evidences show that circular RNA (circRNA) has an important role in ovarian cancer (OC). Hsa_circ_0015326 is a newly discovered upregulated circRNA in OC, but its role and mechanism in OC have not been studied yet.
Methods: Quantitative real-time PCR was used to detect the expression of hsa_circ_0015326, microRNA (miR)-127-3p and MYB. The viability, colony number, cell cycle process, invasion, migration and apoptosis of cells were determined using cell counting kit 8 assay, colony formation assay, flow cytometry, transwell assay and wound healing assay. Moreover, the protein expression levels of metastasis, proliferation, apoptosis markers and MYB were assessed using Western blot analysis. The interaction between miR-127-3p and hsa_circ_0015326 or MYB was confirmed by dual-luciferase reporter assay and RNA immunoprecipitation assay. Xenograft tumors were built to explore the role of hsa_circ_0015326 in OC tumor growth in vivo.
Results: Elevated expression of hsa_circ_0015326 was identified in OC tissues and cells. Loss-of-function experiments suggested that silenced hsa_circ_0015326 inhibited the proliferation, invasion, migration, and promoted the apoptosis of OC cells in vitro, as well as inhibited OC tumorigenesis in vivo. Mechanically, hsa_circ_0015326 sponged miR-127-3p and miR-127-3p targeted MYB. The rescue experiments revealed that miR-127-3p inhibitor reversed the inhibitory effect of hsa_circ_0015326 silencing on OC progression, and MYB overexpression reversed the suppressive effect of miR-127-3p on OC progression. In addition, our data indicated that MYB expression was positively regulated by hsa_circ_0015326.
Conclusion: This study showed that hsa_circ_0015326 could facilitate OC progression by regulating the miR-127-3p/MYB axis, which suggested that it might become a potential target for the treatment of OC.
Keywords: ovarian cancer, hsa_circ_0015326, miR-127-3p, MYB
Introduction
Ovarian cancer (OC) is a common malignant tumor occurring in the ovary.1,2 Because early diagnosis is difficult, OC is mostly diagnosed in the middle and late stages, so OC is a high mortality disease.3,4 OC is often accompanied by metastasis, and the postoperative recurrence rate is relatively high.5,6 In recent years, the emergence of molecular targeted therapy has provided new hope for cancer treatment.7,8 Therefore, it is urgent to clarify the molecular mechanism that affects OC progression and provide new molecular therapeutic targets for the clinical treatment of OC.
Circular RNAs (circRNAs) are a class of non-coding RNAs discovered in recent years.9 Many studies have confirmed that circRNA has great potential value in the diagnosis and treatment of diseases, and it is closely related to the progression of many diseases including cancer.10,11 Importantly, circRNA molecules are rich in microRNA (miRNA) binding sites and can play a regulatory role by acting as a competitive endogenous RNA (ceRNA) for miRNA.12,13 For example, hsa_circ_101996 was confirmed to serve as a sponge of miR-8075 to accelerate cervical cancer proliferation and invasion by regulating TPX2.14 Hsa_circ_0051240 had been shown to be a ceRNA of miR-637 to mediate KLK4 expression, thereby facilitating OC proliferation and metastasis.15 Also, circMTO1 was considered as a biomarker for OC, which could suppress OC proliferation and invasion via regulating miR-182-5p/KLF15.16 Hence, elucidating the role of circRNA in cancers can provide us with a deeper understanding of the tumorigenesis mechanism, thus providing a new direction for cancer treatment.
In previous studies, Gong et al used microarray analysis to screen the differentially expressed circRNA in OC tissues and normal tissues and revealed that hsa_circ_0015326 was obviously upregulated in OC tissues.17 However, the role and underlying mechanism of hsa_circ_0015326 in OC are still unclear. Therefore, we chose hsa_circ_0015326 to explore its role in OC progression. Using bioinformatics analysis, we inferred the miRNA that interacted with hsa_circ_0015326 and predicted the downstream genes of this miRNA, thus perfecting the hypothesis of the circRNA/miRNA/mRNA axis in OC.
Materials and Methods
Clinical Samples
Clinical tissue samples were collected from OC patients (n = 41) and patients with benign gynecological diseases (normal healthy controls, n = 22) undergoing surgery in Yongchuan Hospital of Chongqing Medical University. All patients provided written informed consent. Both OC tumor tissues and normal tissues were frozen and stored in liquid nitrogen until further analysis. The Ethics Committee of Yongchuan Hospital of Chongqing Medical University approved this study. Our study was conducted in accordance with the Declaration of Helsinki.
Cell Culture and Transfection
Human OC cells (A2780 and SKOV3) and normal ovarian epithelial cells (IOSE-80) were purchased from Biovector National Typical Culture Collection (NTCC, Beijing, China) and maintained in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Gibco) and 1% penicillin-streptomycin liquid (Gibco). All cells were preserved in a humidified incubator with 5% CO2 at 37°C.
The small interfering RNA (siRNA) against hsa_circ_0015326 (si-hsa_circ_0015326#1, si-hsa_circ_0015326#2, and si-hsa_circ_0015326#3) and its negative control (si-NC), miR-127-3p mimic or inhibitor and their corresponding controls (mimic NC or inhibitor NC), pcDNA MYB overexpression vector (pcDNA-MYB) and its negative control (pcDNA) were attained from Genechem (Shanghai, China). The siRNAs, mimics, inhibitors and vectors were transfected into A2780 and SKOV3 cells using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA).
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was isolated using RNeasy Mini kit (Qiagen, Duesseldorf, Germany). Then, cDNA was reversely synthesized using SuperScript cDNA Synthesis Kit (Invitrogen). QRT-PCR was performed with SYBR Green Master (Roche, Basel, Switzerland) in PCR system. GAPDH and U6 were used as internal control. The primer sequences were shown as follows: hsa_circ_0015326, F 5ʹ-CTTTACGGGCAATGGCAACC-3ʹ, R 5ʹ-GCCACAGAATCAGATGATCCAC-3ʹ; miR-127-3p, 5ʹ-GGGTCGGATCCGTCTGAGC-3ʹ, R 5ʹ-CAGTGCGTGTCGTGGAGT-3ʹ; miR-194-5p, F 5ʹ-GCGGCGGTGTAACAGCAACTCC-3ʹ, R 5ʹ-ATCCAGTGCAGGGTCCGAGG-3ʹ; miR-515-5p, F 5ʹ-CGGGTTCTCCAAAAGAAAGCA-3ʹ, R 5ʹ-CAGCCACAAAAGAGCACAAT-3ʹ; MYB, F 5ʹ-ACAGATGGGCAGAAATCGCA-3ʹ, R 5ʹ-GCTGGCTGGCTTTTGAAGAC-3ʹ; GAPDH, F 5ʹ-AGCTCACTGGCATGGCCTTC-3ʹ, R 5ʹ-CGCCTGCTTCACCACCTTCT-3ʹ; U6, F 5ʹ-ATACAGAGAAAGTTAGCACGG-3ʹ, R 5ʹ-GGAATGCTTCAAAGAGTTGTG-3ʹ. The 2−ΔΔCT method was used to detect relative expression.
Agarose Gel Electrophoresis
1% agarose gel was prepared using agarose (0.5 g, Beyotime, Shanghai, China) and tris-acetate-EDTA (TAE) buffer (50 mL, Beyotime). 50 bp DNA Ladder (Solarbio, Beijing, China) was used as a DNA marker. The PCR production of hsa_circ_0015326 and the DNA Ladder H1 were added into the agarose gel for electrolysis (110 V, 15 min).
Cell Viability
Transfected A2780 and SKOV3 cells were reseeded onto 96-well plates. At the indicated time points, 10 μL cell counting kit 8 (CCK8) reagent (Roche) was added to each well. After cultured for 4 h, the optical density (OD) value was detected at 450 nm using a microplate reader.
Colony Formation Assay
After transfection, A2780 and SKOV3 cells were seeded in 6-well plates and cultured for 2 weeks. Then, the colonies were fixed with paraformaldehyde (Boster, Wuhan, China) and stained with crystal violet (Beyotime) to count its number under a microscope.
Flow Cytometry
Cell cycle process and cell apoptosis were determined using this assay. A2780 and SKOV3 cells were harvested and washed with cold phosphate buffer saline (PBS, Beyotime). The cell suspensions were fixed with 70% ethanol (Jianxing, Guangzhou, China), incubated with RNase A and propidium iodide (all from Beyotime) to analyze cell cycle process using flow cytometer. Additionally, the cell suspensions were suspended with Annexin V-FITC binding buffer (Beyotime) and then stained with Annexin V-FITC (Beyotime) and propidium iodide to analyze cell apoptosis rate using flow cytometer.
Cell Invasion Assay
A2780 and SKOV3 cells suspended with serum-free medium were added into the transwell upper chambers (Corning Inc., Corning, NY, USA) pre-coated with Matrigel (Corning Inc.). At the same time, serum medium was filled in the lower chambers. After 24 h, the invaded cells were fixed with paraformaldehyde and stained with crystal violet. Three fields were randomly selected for photographing under a microscope (100 ×), and the number of invaded cells was counted.
Wound Healing Assay
A2780 and SKOV3 cells were cultured in 6-well plates. After the cells reached 90% confluences, wounds were scratched using a 20 μL pipette tip. After that, the cells were washed with PBS and then photographed under a microscope (40 ×) to count wound area. After culturing in serum-free medium for 24 h, the wound area of cells was measured and the cell migration ratio was calculated using Image J software (National Institutes of Health, Bethesda, Maryland, USA).
Western Blot (WB)
Protein was extracted using RIPA reagent (Beyotime), and then isolated by SDS-PAGE and transferred to PVDF membranes (Roche). After incubated with non-fat milk for 1 h, the membranes were hatched with primary antibodies against E-cadherin (E-cad, 1:2000, Beyotime), Vimentin (1:5000, Beyotime), CyclinD1 (1:8000, Abcam, Cambridge, MA, USA), Bax (1:1000, Abcam), MYB (1:5000, Abcam), or GAPDH (1:2500, Abcam) overnight at 4°C. Following incubated with HRP-conjugated rabbit IgG (1:50,000, Abcam) for 1 h, protein bands were detected with ECL luminescence reagent (Sangon, Shanghai, China).
Dual-Luciferase Reporter Assay
The wild type (wt) and mutant (mut) sequences of hsa_circ_0015326 or MYB 3ʹUTR were separately cloned to the pmirGLO vectors (Promega, Madison, WI, USA), respectively. 293T cells (NTCC) were seeded in 96-well plates. The constructed vectors were co-transfected with miR-127-3p mimic or mimic NC into 293T cells. After 48 h, luciferase activity was quantified using the Dual-Luciferase Reporter Assay System (Promega).
RNA Immunoprecipitation (RIP) Assay
Magna RIP Kit was obtained from Millipore (Billerica, MA, USA). A2780 and SKOV3 cells were lysed with RIP lysis buffer. The cell lysates were incubated with magnetic bead coupled with the antibodies against Argonaute 2 (anti-Ago2, 1:500, Abcam) and immunoglobulin G (anti-IgG, 1:1000, Abcam). Then, the immunocomplexes were incubated with proteinase K, and then the enrichment of hsa_circ_0015326, miR-127-3p and MYB was measured by qRT-PCR. The lysate not incubated with the antibody was used as Input.
Xenograft Tumors
Male BALB/c-nude mice were bought from Vital River (Beijing, China) and divided into 2 groups (n = 6). SKOV3 cells (5 × 106) transfected with lentivirus short hairpin against hsa_circ_0015326 (sh-hsa_circ_0015326) or its control (sh-NC) were inoculated to the left flank of mice. Tumor length and width were measured every 7 days to count the tumor volume (length × width2/2). After 28 days, mice were sacrificed and tumor tissue was collected and photographed. The paraffin sections were prepared from tumor tissues and incubated with ki-67, E-cad and Vimentin antibodies for immunohistochemical (IHC) staining. All animal studies were conducted according to the rules of the Ethics Committee of Yongchuan Hospital of Chongqing Medical University and were performed in compliance with the ARRIVE guidelines and the Basel Declaration.
Statistical Analysis
The results were evaluated as means ± standard deviation of at least 3 independent experiments. GraphPad Prism 5.0 software (GraphPad Inc., La Jolla, CA, USA) was used for statistical analysis. Differences between groups were performed using one-way analysis of variance or Student’s t-test. Pearson correlation analysis was utilized for correlation analysis. P < 0.05 was considered statistically significant.
Results
Hsa_circ_0015326 Had Increased Expression in OC Tissues and Cells
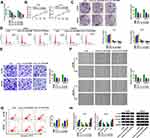
Hsa_circ_0015326 was located on chromosome 1 and was circularized by the three exons of the RABGAP1L gene (Figure 1A). Agarose gel electrophoresis was performed on the PCR amplification product of hsa_circ_0015326, and the results showed that there was a clear band at 177 bp, indicating that the constructed hsa_circ_0015326 primer had good specificity (Figure 1B). In OC tumor tissues, we discovered a markedly elevated expression of hsa_circ_0015326 compared to normal tissues (Figure 1C). According to the median expression of hsa_circ_0015326 in OC tissues, OC tissues were divided into high hsa_circ_0015326 expression group and low hsa_circ_0015326 expression group. By analyzing the relationship between the expression of hsa_circ_0015326 and the clinicopathological characteristics of patients, we found that the high hsa_circ_0015326 expression was related to serum CA-125, tumor size, lymph node metastasis, FIGO stage and tumor grade of patients (Table 1). Moreover, hsa_circ_0015326 also was higher expressed in OC cells (A2780 and SKOV3) than in IOSE-80 cells (Figure 1D). Therefore, we speculated that hsa_circ_0015326 might play an important role in OC progression.
 |
Table 1 Association of Hsa_circ_0015326 Expression with Clinicopathological Characteristics in 41 Patients of Ovarian Cancer |
Downregulation of Hsa_circ_0015326 Repressed the Proliferation, Invasion, Migration, and Enhanced the Apoptosis of OC Cells
For investigating the role of hsa_circ_0015326 in OC, we constructed three siRNAs of hsa_circ_0015326. By detecting hsa_circ_0015326 expression, we determined that all three siRNAs could significantly reduce hsa_circ_0015326 expression, especially with the best effects of si-hsa_circ_0015326#2 and si-hsa_circ_0015326#3 (Figure 2A). Therefore, si-hsa_circ_0015326#2 and si-hsa_circ_0015326#3 were used for functional experiments. CCK8 assay and colony formation assay results suggested hsa_circ_0015326 knockdown could decrease the viability and the numbers of colonies of A2780 and SKOV3 cells (Figure 2B and C). Silenced hsa_circ_0015326 promoted the cell number in the G0/G1 phase and inhibited that of in the S phase, indicated that it could induce cell cycle arrest (Figure 2D). Moreover, the number of invaded cells and cell migration ratio in A2780 and SKOV3 cells also were repressed by hsa_circ_0015326 silencing (Figure 2E and F). The results of flow cytometry showed that downregulated hsa_circ_0015326 could accelerate the apoptosis rate of A2780 and SKOV3 cells (Figure 2G). In addition, we found that the protein levels of E-cad and Bax were markedly increased while the protein levels of Vimentin and CyclinD1 were obviously decreased in the hsa_circ_0015326 silencing groups compared to the control group (Figure 2H). Our data determined that hsa_circ_0015326 had a pro-oncogenic effect in OC.
Hsa_circ_0015326 Served as a Sponge of miR-127-3p
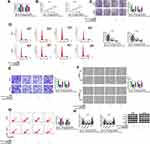
In order to elucidate the molecular mechanism of hsa_circ_0015326, we used the starbase3.0 software and circinteractome software to predict the targeted miRNAs that could interact with hsa_circ_0015326, and the results showed that miR-127-3p, miR-194-5p and miR-515-5p were the targeted miRNAs for hsa_circ_0015326 (Figure 3A). In A2780 and SKOV3 cells with hsa_circ_0015326 knockdown, we found that the expression of all three miRNAs was notably increased, especially the upregulation of miR-127-3p was particularly significant (Figure 3B). Therefore, miR-127-3p was selected as the hsa_circ_0015326 targeted miRNA for validation. In OC tissues and cells, we discovered a remarkably downregulated miR-127-3p compared to the negative controls (Figure 3C and D). Correlation analysis revealed that miR-127-3p expression was negatively correlated with hsa_circ_0015326 expression in OC tissues (Figure 3E). Based on the complementary binding sequence between hsa_circ_0015326 and miR-127-3p, we constructed the hsa_circ_0015326 wt and hsa_circ_0015326 mut vectors (Figure 3F). Dual-luciferase reporter assay results indicated that miR-127-3p overexpression significantly reduced the luciferase activity of hsa_circ_0015326 wt vector, but did not affect the luciferase activity of hsa_circ_0015326 mut vector (Figure 3G). In addition, the results of RIP assay suggested that hsa_circ_0015326 and miR-127-3p were markedly enriched in anti-Ago2 compared to anti-IgG (Figure 3H), indicating that there was an interaction between the two. By detecting the expression of miR-127-3p, we determined that miR-127-3p inhibitor and mimic had good inhibitory and promotion effects on miR-127-3p expression in A2780 and SKOV3 cells (Figure 3I), so further studies could be carried out.
Hsa_circ_0015326 Regulated OC Progression by Sponging miR-127-3p
To further confirm that whether hsa_circ_0015326 sponged miR-127-3p to regulate OC progression, si-hsa_circ_0015326#3 and miR-127-3p inhibitor were co-transfected into A2780 and SKOV3 cells to perform the rescue experiments. MiR-127-3p inhibitor significantly reduced the miR-127-3p expression promoted by hsa_circ_0015326 silencing, showing that the transfection of both was successful (Figure 4A). As exhibited in Figure 4B–D, it was observed that the inhibitory effect of hsa_circ_0015326 knockdown on the viability, colony number and cell cycle process of A2780 and SKOV3 cells could be reversed by the addition of miR-127-3p inhibitor. Also, miR-127-3p inhibitor reversed the suppressive effect of hsa_circ_0015326 silencing on the invaded cell number and the migration ratio, as well as the promotion effect on the apoptosis rate of A2780 and SKOV3 cells (Figure 4E–G). Additionally, the enhancing effect of hsa_circ_0015326 knockdown on E-cad and Bax protein levels and the reducing effect on Vimentin and CyclinD1 protein levels also were inverted by miR-127-3p inhibitor in A2780 and SKOV3 cells (Figure 4H). These results illuminated that miR-127-3p was involved in the regulation of hsa_circ_0015326 on OC progression.
MYB Was a Target of miR-127-3p
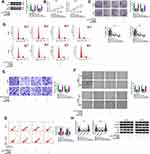
To find the downstream targets of miR-127-3p, we used the starbase3.0 software to make the bioinformatics predictions. We found that the 3ʹUTR of MYB contained the complementary binding sites with miR-127-3p (Figure 5A). Further analysis revealed that the luciferase activity of MYB 3ʹUTR wt vector instead of MYB 3ʹUTR mut vector could be significantly reduced by miR-127-3p overexpression (Figure 5B). In addition, we also discovered significantly enrichment of MYB and miR-127-3p in anti-Ago2 (Figure 5C). Furthermore, the expression of MYB in OC tissues and cells was also detected. The results showed that MYB was remarkably overexpressed in OC tumor tissues and cells compared with the control group (Figure 5D–F), and its mRNA expression in OC tumor tissues was negatively correlated with miR-127-3p expression (Figure 5G). For further research, we constructed the pcDNA MYB overexpression vector. WB analysis results showed that MYB protein expression was obviously increased in A2780 and SKOV3 cells after transferred with pcDNA MYB overexpression vector (Figure 5H).
Overexpressed MYB Reversed the Inhibitory of miR-127-3p on OC Progression
Then, miR-127-3p mimic and pcDNA MYB overexpression vector were co-transfected into A2780 and SKOV3 cells. Our results showed that miR-127-3p mimic significantly inhibited MYB protein expression, but pcDNA MYB overexpression vector could reverse this effect (Figure 6A). Further experiments indicated that miR-127-3p suppressed cell viability, colony number, and induced cell cycle arrest in A2780 and SKOV3 cells, while this effect could be recovered by MYB overexpression (Figure 6B–D). Meanwhile, overexpressed MYB also reversed the decreasing effect of miR-127-3p on the invaded cell number, migration ratio, and the increasing effect on the apoptosis rate of A2780 and SKOV3 cells (Figure 6E–G). Furthermore, miR-127-3p overexpression accelerated the protein levels of E-cad and Bax and hindered the protein levels of Vimentin and CyclinD1 in A2780 and SKOV3 cells, and these effects also could be reversed by MYB overexpression (Figure 6H). Our results manifested that miR-127-3p regulated OC progression by targeting MYB.
MYB Expression Was Positively Regulated by Hsa_circ_0015326
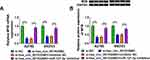
In order to further confirm the speculation of the hsa_circ_0015326/miR-127-3p/MYB axis, we investigated the regulation of hsa_circ_0015326 on MYB expression. We found that hsa_circ_0015326 knockdown remarkably decreased MYB expression, and this effect could be eliminated by miR-127-3p inhibitor (Figure 7A). The detection result of MYB protein level was consistent with the mRNA level (Figure 7B). Hence, we proposed that hsa_circ_0015326 indirectly regulated MYB expression by targeting miR-127-3p in OC.
Hsa_circ_0015326 Knockdown Inhibited OC Tumorigenesis in vivo
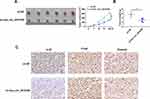
To further confirm the positive role of hsa_circ_0015326 in OC, we performed constructed xenograft tumors by performing in vivo experiments. Our results showed that the tumor size and volume were markedly reduced after inhibiting hsa_circ_0015326 expression (Figure 8A). Also, the tumor weight in the sh-circ_0015326 group also was decreased compared to the sh-NC group (Figure 8B). IHC staining showed that the ki-67 and Vimentin positive cells were repressed while the E-cad positive cells were enhanced in the sh-circ_0015326 group (Figure 8C). These data confirmed that hsa_circ_0015326 also could promote the tumorigenesis of OC.
Discussion
Numerous studies have found that circRNA is abnormally expressed between cancer and normal tissues, suggesting that circRNA may be a key factor in cancer progression.10,11 CircRNA has been proven to be a biomarker for the malignant progression of cancer, including gastric cancer,18 lung adenocarcinoma,19 and pancreatic cancer.20 Many circRNAs have been proven to be biomarkers and therapeutic targets for OC, such as circRHOBTB3,21 circ-ITCH,22 and circ-UBAP2.23 Here, we explored the role of a newly discovered circRNA, hsa_circ_0015326, in OC development. Our results showed that hsa_circ_0015326 was highly expressed in OC tissues and cells, which was consistent with the microarray analysis results of Gong et al.17 Loss-of-function experiments revealed that hsa_circ_0015326 knockdown could repress proliferation, invasion, migration, and accelerate apoptosis in OC cells. Also, animal experiments showed that silenced hsa_circ_0015326 also suppressed OC tumorigenesis. These results suggested that hsa_circ_0015326 had a pro-cancer role in OC. The elucidation of hsa_circ_0015326 function provided a new potential target for the molecular therapy of OC.
CircRNAs can be used as miRNA sponges have been confirmed by many studies.12,13 Using bioinformatics analysis, we found that hsa_circ_0015326 interacted with miR-127-3p in OC. MiR-127-3p is low expressed in many cancers and has been found to act as a tumor suppressor to regulate cancer progression. For example, miR-127-3p was downregulated in oral squamous cell carcinoma, which could inhibit cancer proliferation and metastasis via targeting KIF3B.24 MiR-127-3p was found to be a significantly underexpressed miRNA in epithelial OC, so it could be used as a biomarker for epithelial OC.25 Bi et al suggested that overexpressed miR-127-3p targeted BAG5 to restrain epithelial OC proliferation and invasion.26 Also, Xia et al indicated that miR-127 played an anti-cancer role in OC.27 Similar to the previous results, we confirmed that miR-127-3p expression in OC was obviously decreased. The rescue experiments illuminated that the anti-proliferation, anti-metastasis and pro-apoptosis of hsa_circ_0015326 silencing on OC could be abolished by miR-127-3p inhibitor, confirming that hsa_circ_0015326 targeted miR-127-3p to regulate the progression of OC. In addition, the tumor-suppressive effect of miR-127-3p also helped us better understand the positive role of hsa_circ_0015326 in OC.
MYB gene family is an important transcription factor family, which plays a vital role in regulating cell survival, differentiation and cycle.28,29 In recent years, many studies have also found that MYB can regulate cancer progression.30,31 Zhang et al showed that MYB was highly expressed in OC and could promote epithelial–mesenchymal transition and cisplatin resistance in OC.32 Furthermore, previous studies also indicated that MYB overexpression could promote OC proliferation, invasion and cisplatin resistance by activating NF-κB/STAT3 pathway.33 In our study, we proposed that MYB was targeted by miR-127-3p, and its expression was overexpressed in OC tissues and cells. In addition, the reversal effect of MYB on the suppressive effect of miR-127-3p indicated that MYB was a key regulator to promote OC progression. Moreover, we also confirmed that hsa_circ_0015326 positively regulated MYB expression by sponging miR-127-3p.
In summary, we found that hsa_circ_0015326 was closely related to OC progression. Functional and mechanism analysis suggested that hsa_circ_0015326 enhanced the proliferation, metastasis and apoptosis inhibition of OC, which was achieved by regulating the miR-127-3p/MYB axis. In general, our findings proposed that hsa_circ_0015326 might be a potential new biomarker for OC treatment.
Ethics Approval
Our study was approved by the Institutional Review Board in Yongchuan Hospital of Chongqing Medical University.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
There is no funding to report.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Momenimovahed Z, Tiznobaik A, Taheri S, et al. Ovarian cancer in the world: epidemiology and risk factors. Int J Womens Health. 2019;11:287–299. doi:10.2147/IJWH.S197604
2. La Vecchia C. Ovarian cancer: epidemiology and risk factors. Eur J Cancer Prev. 2017;26(1):55–62. doi:10.1097/CEJ.0000000000000217
3. Malvezzi M, Carioli G, Rodriguez T, et al. Global trends and predictions in ovarian cancer mortality. Ann Oncol. 2016;27(11):2017–2025. doi:10.1093/annonc/mdw306
4. Dinkelspiel HE, Champer M, Hou J, et al. Long-term mortality among women with epithelial ovarian cancer. Gynecol Oncol. 2015;138(2):421–428. doi:10.1016/j.ygyno.2015.06.005
5. Jayson GC, Kohn EC, Kitchener HC, et al. Ovarian cancer. Lancet. 2014;384(9951):1376–1388. doi:10.1016/S0140-6736(13)62146-7
6. Wang Z, Zhao D, Liu R, et al. Ovarian cancer metastasis to the breast 18 years after the initial diagnosis: a case report. Medicine (Baltimore). 2019;98(43):e17577. doi:10.1097/MD.0000000000017577
7. Guan LY, Lu Y. New developments in molecular targeted therapy of ovarian cancer. Discov Med. 2018;26(144):219–229.
8. Lee YT, Tan YJ, Oon CE. Molecular targeted therapy: treating cancer with specificity. Eur J Pharmacol. 2018;834:188–196. doi:10.1016/j.ejphar.2018.07.034
9. Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32(5):309–316. doi:10.1016/j.tig.2016.03.002
10. Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267–274. doi:10.1016/j.ebiom.2018.07.036
11. Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev. 2018;48:121–127. doi:10.1016/j.gde.2017.11.007
12. Jiang WD, Yuan PC, Wang E. Molecular network-based identification of competing endogenous RNAs in bladder cancer. PLoS One. 2019;14(8):e0220118. doi:10.1371/journal.pone.0220118
13. Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79. doi:10.1186/s12943-018-0827-8
14. Song T, Xu A, Zhang Z, et al. CircRNA hsa_circRNA_101996 increases cervical cancer proliferation and invasion through activating TPX2 expression by restraining miR-8075. J Cell Physiol. 2019;234(8):14296–14305. doi:10.1002/jcp.28128
15. Zhang M, Xia B, Xu Y, et al. Circular RNA (hsa_circ_0051240) promotes cell proliferation, migration and invasion in ovarian cancer through miR-637/KLK4 axis. Artif Cells Nanomed Biotechnol. 2019;47(1):1224–1233. doi:10.1080/21691401.2019.1593999
16. Wang N, Cao QX, Tian J, et al. Circular RNA MTO1 inhibits the proliferation and invasion of ovarian cancer cells through the miR-182-5p/KLF15 axis. Cell Transplant. 2020;29:963689720943613. doi:10.1177/0963689720943613
17. Gong J, Xu X, Zhang X, et al. Circular RNA-9119 suppresses in ovarian cancer cell viability via targeting the microRNA-21-5p-PTEN-Akt pathway. Aging (Albany NY). 2020;12(14):14314–14328. doi:10.18632/aging.103470
18. Tang W, Fu K, Sun H, et al. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer. 2018;17(1):137. doi:10.1186/s12943-018-0888-8
19. Zhu X, Wang X, Wei S, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284(14):2170–2182. doi:10.1111/febs.14132
20. Yang F, Liu DY, Guo JT, et al. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol. 2017;23(47):8345–8354. doi:10.3748/wjg.v23.i47.8345
21. Yalan S, Yanfang L, He C, et al. Circular RNA circRHOBTB3 inhibits ovarian cancer progression through PI3K/AKT signaling pathway. Panminerva Med. 2020. doi:10.23736/S0031-0808.20.03957-9
22. Lin C, Xu X, Yang Q, et al. Circular RNA ITCH suppresses proliferation, invasion, and glycolysis of ovarian cancer cells by up-regulating CDH1 via sponging miR-106a. Cancer Cell Int. 2020;20(1):336. doi:10.1186/s12935-020-01420-7
23. Xu Q, Deng B, Li M, et al. circRNA-UBAP2 promotes the proliferation and inhibits apoptosis of ovarian cancer though miR-382-5p/PRPF8 axis. J Ovarian Res. 2020;13(1):81. doi:10.1186/s13048-020-00685-w
24. Ji L, Zhu ZN, He CJ, et al. MiR-127-3p targets KIF3B to inhibit the development of oral squamous cell carcinoma. Eur Rev Med Pharmacol Sci. 2019;23(2):630–640. doi:10.26355/eurrev_201901_16877
25. Rattanapan Y, Korkiatsakul V, Kongruang A, et al. MicroRNA expression profiling of epithelial ovarian cancer identifies new markers of tumor subtype. Microrna. 2020;9(4):289–294. doi:10.2174/2211536609666200722125737
26. Bi L, Yang Q, Yuan J, et al. MicroRNA-127-3p acts as a tumor suppressor in epithelial ovarian cancer by regulating the BAG5 gene. Oncol Rep. 2016;36(5):2563–2570. doi:10.3892/or.2016.5055
27. Liu X, Meng Z, Xing Y, et al. MiR-127 inhibits ovarian cancer migration and invasion by up-regulating ITGA6. Minerva Med. 2019. doi:10.23736/S0026-4806.19.06237-2
28. Wang X, Angelis N, Thein SL. MYB - A regulatory factor in hematopoiesis. Gene. 2018;665:6–17. doi:10.1016/j.gene.2018.04.065
29. Dubos C, Stracke R, Grotewold E, et al. MYB transcription factors in arabidopsis. Trends Plant Sci. 2010;15(10):573–581. doi:10.1016/j.tplants.2010.06.005
30. Yang RM, Nanayakkara D, Kalimutho M, et al. MYB regulates the DNA damage response and components of the homology-directed repair pathway in human estrogen receptor-positive breast cancer cells. Oncogene. 2019;38(26):5239–5249. doi:10.1038/s41388-019-0789-3
31. Xu LH, Zhao F, Yang WW, et al. MYB promotes the growth and metastasis of salivary adenoid cystic carcinoma. Int J Oncol. 2019;54(5):1579–1590. doi:10.3892/ijo.2019.4754
32. Zhang XY, Li YF, Ma H, et al. Regulation of MYB mediated cisplatin resistance of ovarian cancer cells involves miR-21-wnt signaling axis. Sci Rep. 2020;10(1):6893. doi:10.1038/s41598-020-63396-8
33. Tian M, Tian D, Qiao X, et al. Modulation of Myb-induced NF-kB -STAT3 signaling and resulting cisplatin resistance in ovarian cancer by dietary factors. J Cell Physiol. 2019;234(11):21126–21134. doi:10.1002/jcp.28715
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.