Back to Journals » Cancer Management and Research » Volume 12
hsa_circ_0008285 Facilitates the Progression of Cervical Cancer by Targeting miR-211-5p/SOX4 Axis
Received 31 December 2019
Accepted for publication 26 April 2020
Published 26 May 2020 Volume 2020:12 Pages 3927—3936
DOI https://doi.org/10.2147/CMAR.S244317
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Beicheng Sun
Youpeng Bai, Xicong Li
Obstetrical Department, Luoyang Central Hospital Affiliated to Zhengzhou University, Luoyang 471009, Henan, People’s Republic of China
Correspondence: Youpeng Bai Email [email protected]
Introduction: Emerging evidence has demonstrated that circRNAs are implicated in the progression of cervical cancer (CC). However, the roles and underlying mechanisms of circRNAs remain unclear in CC.
Methods: QRT-PCR was performed to detect hsa_circ_0008285 expression in CC tissues and cell lines. The roles of hsa_circ_0008285 on CC progression were explored by function assays. Next, the underlying mechanisms of hsa_circ_0008285 in CC progression were determined by bioinformatics analysis, dual-luciferase reporter and RIP assays.
Results: In the present study, we identified a new circRNA hsa_circ_0008285, which was significantly up-regulated in CC tissues and cell lines. Loss-of-function assays showed that hsa_circ_0008285 suppression reduced the proliferation and invasion of CC cells in vitro and reduced tumor growth in vivo. In mechanism, bioinformatics analysis, dual-luciferase reporter and RIP assays showed that hsa_circ_0008285 served as a sponge for miR-211-5p in CC. Next, we confirmed that SOX4 served as a target gene for miR-211-5p in CC. Additionally, we revealed that miR-211-5p inhibitors abolished the effects of hsa_circ_0008285 on SOX4 expression in CC cells.
Conclusion: Therefore, our research highlighted that hsa_circ_0008285 promoted CC progression via serving as a ceRNA of miR-211-5p to release SOX4, which might provide a potential therapeutic target for tumor treatment.
Keywords: cervical cancer, hsa_circ_0008285, miR-211-5p, SOX4, ceRNA
Introduction
Cervical cancer (CC) is the second highest incidence and the fourth dominating cause of cancer-induced death in women worldwide.1,2 Human papillomavirus (HPV) is the major cause of CC, and often related to risk sexual behaviors, early age at first intercourse, early beginning of sexual activities, multiple pregnancies with Chlamydia association and immunosuppression.3,4 Although the therapeutic approaches for CC have been greatly improved over the past decades, the 5-year overall survival rate remains poor due to metastases and/or relapse.5,6 Therefore, it is necessary to explore potential molecular mechanisms of CC progression, which might offer new therapeutic targets and approaches for CC patients.
With the development of next generation sequencing and bioinformatics algorithm, the abundance of non-coding RNAs (ncRNAs) are implicated in various developmental stages in mammal and pathological conditions.7,8 Among these non-coding transcripts, circular RNAs (circRNAs) represent a novel class of endogenous ncRNAs and generate by back-splicing with covalently closed-loop structures, which endow them with a higher stabilization.9,10 Growing evidence suggested that circRNAs play important roles in the pathological development of human diseases, including cancer. For example, Ge et al found that circMTO1 suppressed colorectal cancer cells proliferation and invasion through regulating Wnt/beta-catenin pathway.11 Xue et al showed that hsa_circ_0081143 promoted cisplatin resistance through regulating miR-646-CDK6 axis in gastric cancer.12 Zhou et al found that circPCNXL2 sponged miR-153 to promote renal cancer cells proliferation and invasion by regulating ZEB2 expression.13 However, the functions and molecular mechanisms of circRNAs in tumorigenesis remain unclear.
In the present study, by analyzing the GSE102686 chip, we found hsa_circ_0008285 was one of the most upregulated circRNAs in CC. Next, we confirmed that hsa_circ_0008285 expression was highly expressed in CC tissues and cell lines. High hsa_circ_0008285 expression promoted CC cells proliferation and invasion abilities. In mechanism, hsa_circ_0008285 might exert as an oncogenic circRNA in CC progression through regulating the miR-211-5p/SOX4 axis. These findings suggested that hsa_circ_0008285 might serve as a potential therapeutic target for CC treatment.
Materials and Methods
Tissues Samples
A total of 37 pairs CC tissues were provided by Luoyang Central Hospital Affiliated to Zhengzhou University between May 2017 and May 2018. All tissue samples were immediately snap-frozen in liquid nitrogen after surgery and stored at −80°C until RNA extraction. All patients did not receive radiation or chemotherapy treatment before surgery. The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of the Luoyang Central Hospital Affiliated to Zhengzhou University. Written informed consent was received from all patients before this study.
Cell Culture and Transfection
Five human CC cell lines (Hela, C33A, SiHa, SW756, and Caski) and human cervical epithelial immortalized cell line (H8) were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China) and incubated in Dulbecco’s modified Eagle’s medium (DMEM) added 10% fetal bovine serum (FBS; Invitrogen, MA, USA), 100 U/mL of penicillin and 100 mg/mL of streptomycin at 37°C with 5% CO2 in a humidified atmosphere.
The siRNA sequences targeting hsa_circ_0008285 (si-circRNA-1 sequence is 5′- ACGGGAAAGGTTGAAAGGATT-3′; si-circRNA-2 sequence is 5′- GGGAAAGGTTGAAAGGATTGT-3′; si-circRNA-3 sequence is 5′- AACGGGAAAGGTTGAAAGGAT-3′), miR-211-5p mimics and inhibitors and corresponding negative controls were obtained from Ribobio (Guangzhou, China). Cells were transfected with these siRNAs or plasmids by using lipofectamine 2000 (Invitrogen, USA) following the instructions.
Real-Time Quantitative PCR
Total RNAs were isolated from patient specimens and cultured cells using TRIzol reagents following the manufacturer’s protocols. 500 ng RNA was reversed transcribed into cDNA using a reverse transcription kit (Takara, Dalian, China). 20 ng RNA was reversed transcribed into cDNA with TaqMan™ Reverse Transcription reagents (Thermo Fisher Scientific, Wilmington, DE). QRT-PCR was performed on an ABI7900 system (Applied Biosystems, CA, USA). The expression of hsa_circ_0008285, miR-211-5p and SOX4 was normalized to GAPDH or U6, respectively. Relative gene expression was analyzed by the 2−ΔΔCt method.
Cell Proliferation Assay
Using Cell Counting Kit‐8 (CCK‐8; Beyotime, Beijing, China) to measure the cell viability. Transfected cells were adjusted to 1 × 105 cells/well and seeded in 6‐well plates. After 24, 48, 72 and 96 h, 20 μL of CCK‐8 solution was added per well. After 4 h, cell proliferation was determined by measuring the optical density (OD) value at 450 nm.
Colony Formation Assay
Transfected cells were plated in 6‐well plates at a density of 1000 cells per well. 2 weeks later, cells were washed by PBS for twice and fixed by methanol for 30 min. The colonies were then stained by 0.1% crystal violet and the numbers of colonies were manually calculated.
Transwell Assay
Transwell assay was performed to assess cell invasion ability. The upper chambers were coated with Matrigel and incubated overnight before the cells were plated. Then transfected cells were cultured in upper chambers with serum‐free medium and DMEM with 10% FBS was placed into the lower chamber. After 24 h, remained cells were carefully wiped away. Finally, the invaded cells were stained with 0.1% crystal violet. The number of invaded cells was counted under a light microscope (Olympus, Tokyo, Japan).
Tumorigenesis Assay
The animal experiments were approved by the Institutional Animal Ethics Committee of the Luoyang Central Hospital Affiliated to Zhengzhou University. All the procedures of animal experiments were performed in strict accordance with the Laboratory animal protection, welfare and ethics rules of the Institutional Animal Ethics Committee of the Luoyang Central Hospital Affiliated to Zhengzhou University.
6 female nude mice (athymic BALB/c) aged 4–6 weeks and weighing 16–20g were housed in pathogen-free conditions. The animals were randomly split into 2 groups (the sh-NC and sh-circ_0008285 groups). 200μL of 1×107 cells were subcutaneously injected into the back flanks of the animals. Tumors was measured each week after the tumor became visible and tumor volume calculated following with the formula: volume = (0.5 × length × width2). After 6 weeks following implantation, the mice were sacrificed and the tumors removed, weighed and photographed.
Dual-Luciferase Reporter Assay
The wild-type (Wt) or mutant (Mut) sequences of hsa_circ_0008285 or SOX4 3ʹUTR containing the putative miR-211-5p binding sites were cloned into the pmirGLO vector, respectively. Cells were co-transfected with pmirGLO-circ_0008285-Wt/Mut or pmirGLO-SOX4-Wt/Mut and miR-211-5p mimics and incubated for another 48 h. The luciferase activity of each group was detected using the Dual-luciferase Reporter Assay System (Promega, Madison, WI, USA).
RNA Immunoprecipitation (RIP)
RIP experiment was performed using the Magna RIP RNA-binding protein immunoprecipitation kit (Millipore) and Ago2 antibody according to the protocol and previous studies.12,13 Co-precipitated RNAs were analyzed using the qRT–PCR analysis.
Statistical Analysis
Statistical analysis was performed using IBM SPSS Statistics Version 19.0 (Chicago, IL, USA). All data were manifested as the means ± standard deviation (SD) from 3 independent experiments. Student’s t-test or ANOVA was used for comparisons between groups. P<0.05 was considered statistically significant.
Results
hsa_circ_0008285 Is Highly Expressed in CC Tissues and Cell Lines
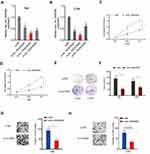
To explore the roles of circRNA in CC progression, we analyzed the GSE102686 chip, and found hsa_circ_0008285 was one of the most upregulated circRNAs in CC (Figure 1A). hsa_circ_0008285 is located in chr6:4891946–4892613, which is spliced from the CDYL gene, and the ultimate length is 667 nt (Figure 1B). Next, we validated the hsa_circ_0008285 expression in CC tissues, qRT-PCR showed hsa_circ_0008285 expression in CC tissues was significantly increased compared with adjacent non-tumor tissues (Figure 1C and D). High hsa_circ_008285 expression was correlated with advanced FIGO stage, and lymph node metastasis in CC patients (Figure 1E and F). In addition, the hsa_circ_0008285 expression levels in CC cell lines (Hela, C33A, SiHa, SW756, and Caski) were higher than that in normal cell line (H8) (Figure 1G). The Hela and C33A cell lines were chosen for further experiments on account of relatively high expression of hsa_circ_0008285.
hsa_circ_0008285 Promotes Proliferation and Invasion of CC Cells
To explore the effects of hsa_circ_0008285 in CC, si-circRNAs were transfected into Hela and C33A cells. The knockdown efficiency was determined by qRT-PCR (Figure 2A and B). CCK-8 assay showed that the proliferation was obviously inhibited by si-circ_0008285 in Hela and C33A cells (Figure 2C and D). Colony formation assay revealed that hsa_circ_0008285 suppression significantly inhibited colony numbers of Hela and C33A cells (Figure 2E and F). Additionally, Transwell assay showed si-circRNA significantly inhibited the invasion abilities in Hela and C33A cells compared to si-NC group (Figure 2G and H). These data indicated that hsa_circ_0008285 might act as an oncogenic circRNA in CC progression.
hsa_circ_0008285 Is Negatively Correlated with miR-211-5p
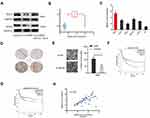
Next, we used the subcellular fractionation assay to explore the location of hsa_circ_0008285 in CC cells. Results showed that hsa_circ_0008285 was mostly scattered in the cytoplasm (Figure 3A). To further explore the underlying mechanism of hsa_circ_0008285 on CC progression, miRanda and RNAhybrid were used to screen miRNA that complementary base pairing with hsa_circ_0008285 (Figure 3B). We found that miR-211-5p is one of the potential target genes of hsa_circ_0008285 with a high score (Figure 3C and D). Luciferase reporter assay verified that miR-211-5p binds directly to hsa_circ_0008285 (Figure 3E). In addition, qRT-PCR showed that hsa_circ_0008285 inhibition dramatically increased the expression of miR-211-5p in Hela and C33A cells (Figure 3F). Besides, miR-211-5p expression in CC tissues was obviously lower than that in adjacent non-tumor tissues (Figure 3G). The expression of hsa_circ_0008285 was appeared to be negatively correlated with miR-211-5p in CC tissues (Figure 3H). Therefore, these data suggested that hsa_circ_0008285 might exert function under the ceRNA pattern by sponging miR-211-5p in CC cells.
hsa_circ_0008285 Positively Regulates SOX4 Expression via miR-211-5p
Next, we explored the downstream target genes for miR-211-5p. Results showed SOX4 was a putative target gene of miR-211-5p with a high score (Figure 4A–C). Dual-luciferase reporter assay confirmed that miR-211-5p binds to SOX4 (Figure 4D). Western blot indicated that miR-211-5p mimics significantly inhibited SOX4 protein expression in Hela and C33A cells (Figure 4E and F). In addition, miR-211-5p expression was appeared to be negatively correlated with SOX4 expression in CC tissues (Figure 4G). These results suggested that SOX4 is one of the target genes of miR-211-5p in CC.
In order to confirm the relationship between hsa_circ_0008285 and SOX4, the expression of SOX4 protein was measured after transfection of si-circRNA. Results showed that SOX4 protein expression was reduced by silencing of hsa_circ_0008285 in Hela and C33A cells, while miR-211-5p inhibitors abolished the effects (Figure 5A). Additionally, qRT-PCR showed that SOX4 mRNA expression was upregulated in CC tissues and cell lines (Figure 5B and C). IHC results further confirmed SOX4 was upregulated in CC tissues (Figure 5D). Transwell invasion assay showed that SOX4 inhibition significantly reduced CC cells invasion ability (Figure 5E). Kaplan-Meier showed that high SOX4 expression was associated with poor OS in CC patients (Figure 5F and G). In addition, the correlation analysis showed that hsa_circ_0008285 expression was positively correlated with SOX4 expression in CC tissues (Figure 5H).
hsa_circ_0008285 Knockdown Inhibits CC Growth in vivo
We further explored the function of hsa_circ_0008285 in vivo. The knockdown efficiency of sh-circRNA in Hela cells was assessed by qRT-PCR (Figure 6A). Then, transfected CC cells were injected into nude mice. Results showed a significantly decreased tumor volume and weights in sh-circRNA group after inoculation 7 weeks (Figure 6B and C). Furthermore, qRT-PCR showed that sh-circRNA significantly reduced hsa_circ_0008285 and SOX4 expression in nude mice tissues, while increased miR-211-5p expression (Figure 6D). These results confirmed that knockdown of hsa_circ_0008285 inhibited CC growth by regulating the miR-211-5p/SOX4 axis (Figure 6E).
Discussion
Increasing studies suggested that circRNAs play critical roles in CC tumorigenesis10 For example, Song et al showed that hsa_circRNA_101996 increased CC cells proliferation and invasion through activating TPX2 expression by restraining miR‐8075.14 Mao et al revealed that circEIF4G2 promoted the malignant features of CC through regulating the miR‑218/HOXA1 axis.15 Tian et al found that circSMARCA5 inhibited the proliferation and invasion in CC by regulating miR-620.16 In the current study, we analyzed the GSE102686 chip and found hsa_circ_0008285 was one of the most upregulated circRNAs, which was further explored in CC tissues and cell lines. Next, function assays revealed that silencing hsa_circ_0008285 significantly reduced the proliferation and invasion ability of CC cells in vitro and reduced tumor growth in vivo. These data suggested that circRNA hsa_circ_0008285 might serve as a new oncogenic circRNA in CC progression.
MicroRNAs (miRNAs) are short ncRNA that participated in pathological processes of cancer.17,18 Increasing studies showed that circRNAs acted as ceRNAs in the regulatory network of circRNA, miRNA and target gene to regulate the expression of their targets.19 The interaction between miRNAs and circRNAs is indispensable in the growth and metastasis of cancer cells.20,21 In the present study, we confirmed that miR-211-5p is one of the target miRNAs of hsa_circ_0008285. Lots of studies showed that miR-211-5p was abnormally expressed in cancer. For example, Wang et al showed that miR-211-5p suppressed renal cancer metastasis abilities by regulating SNAI1.22 Zhang et al found that lncRNA KCNQ1OT1 regulated tongue cancer cells proliferation and cisplatin resistance via miR-211-5p/Ezrin/Fak/Src axis.23 In line with those above studies, our results showed that miR-211-5p was downregulated and _circ_0008285 interacted with miR-211-5p as a competing endogenous RNA to inhibit their action in CC. Collectively, we concluded that hsa_circ_0008285 might exert an oncogenic role by sponging miR-211-5p in CC progression.
SOX4, a member of the C subgroup of SRY-related HMG box (SOX) transcription factor family, which is a crucial transcriptional factor associated with a series of development procedures, including tumorigenesis.24,25 For example, Ruan et al found that overexpression of SOX4 promoted cell migration and invasion of renal cell carcinoma by inducing epithelial-mesenchymal transition.26 Li et al found that miR-132 reduced lung cancer cell progression by regulating SOX4 expression.27 Ding et al showed that circ-DONSON promoted gastric cancer growth and invasion by recruiting the NURF complex to initiate SOX4 expression.28 In the present study, SOX4 was highly expressed in CC tissues and cell lines. Additionally, SOX4 is the direct target of miR-211-5p and positively regulated by hsa_circ_0008285 in CC. These data indicated that hsa_circ_0008285 might positively regulate SOX4 by down-regulating miR-211-5p expression.
In conclusion, our study revealed an oncogenic role of hsa_circ_0008285 in CC progression. Inhibition of hsa_circ_0008285 decreased SOX4 expression by inducing miR-211-5p expression, thereby promoting the proliferation and invasion of CC. Therefore, we suggested that hsa_circ_0008285 might act as a potential therapeutic target for CC treatment.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Franco EL, Duarte-Franco E, Ferenczy A. Cervical cancer: epidemiology, prevention and the role of human papillomavirus infection. CMAJ. 2001;164(7):1017–1025.
3. Sankaranarayanan R, Nene BM, Shastri SS, et al. HPV screening for cervical cancer in rural India. N Engl J Med. 2009;360(14):1385–1394. doi:10.1056/NEJMoa0808516
4. de Martel C, Plummer M, Vignat J, et al. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int J Cancer. 2017;141(4):664–670. doi:10.1002/ijc.30716
5. Clark LH, Jackson AL, Soo AE, et al. Extremes in body mass index affect overall survival in women with cervical cancer. Gynecol Oncol. 2016;141(3):497–500. doi:10.1016/j.ygyno.2016.03.035
6. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
7. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5. doi:10.1038/nrc.2017.99
8. Beermann J, Piccoli M-T, Viereck J, et al. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–1325. doi:10.1152/physrev.00041.2015
9. Pamudurti NR, Bartok O, Jens M, et al. Translation of circRNAs. Mol Cell. 2017;66(1):9–21. e7. doi:10.1016/j.molcel.2017.02.021
10. Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev. 2018;48:121–127. doi:10.1016/j.gde.2017.11.007
11. Ge Z, Li LF, Wang CY, et al. CircMTO1 inhibits cell proliferation and invasion by regulating Wnt/beta-catenin signaling pathway in colorectal cancer. Eur Rev Med Pharmacol Sci. 2018;22(23):8203–8209. doi:10.26355/eurrev_201812_16513
12. Xue M, Li G, Fang X, et al. hsa_circ_0081143 promotes cisplatin resistance in gastric cancer by targeting miR-646/CDK6 pathway. Cancer Cell Int. 2019;19(1):25. doi:10.1186/s12935-019-0737-x
13. Zhou B, Zheng P, Li Z, et al. CircPCNXL2 sponges miR-153 to promote the proliferation and invasion of renal cancer cells through upregulating ZEB2. Cell Cycle. 2018;17(23):2644–2654. doi:10.1080/15384101.2018.1553354
14. Song T, Xu A, Zhang Z, et al. CircRNA hsa_circRNA_101996 increases cervical cancer proliferation and invasion through activating TPX2 expression by restraining miR‐8075. J Cell Physiol. 2019;234(8):14296–14305. doi:10.1002/jcp.28128
15. Mao Y, Zhang L, Li Y. circEIF4G2 modulates the malignant features of cervical cancer via the miR-218/HOXA1 pathway. Mol Med Rep. 2019;19(5):3714–3722. doi:10.3892/mmr.2019.10032
16. Tian JDC, Liang L. Involvement of circular RNA SMARCA5/microRNA-620 axis in the regulation of cervical cancer cell proliferation, invasion and migration. Eur Rev Med Pharmacol Sci. 2018;22:8589–8598. doi:10.26355/eurrev_201812_16622
17. Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015;15(1):38. doi:10.1186/s12935-015-0185-1
18. Eddy SR. Non–coding RNA genes and the modern RNA world. Nat Rev Genet. 2001;2(12):919. doi:10.1038/35103511
19. Qi X, Zhang D-H, Wu N, et al. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718. doi:10.1136/jmedgenet-2015-103334
20. Zhou R, Wu Y, Wang W, et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134–142. doi:10.1016/j.canlet.2018.03.035
21. Zhong L, Wang Y, Cheng Y, et al. Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway. Biochem Biophys Res Commun. 2018;499(4):1044–1049. doi:10.1016/j.bbrc.2018.03.221
22. Wang K, Jin W, Jin P, et al. miR-211-5p suppresses metastatic behavior by targeting SNAI1 in renal cancer. Mol Cancer Res. 2017;15(4):448–456. doi:10.1158/1541-7786.MCR-16-0288
23. Zhang S, Ma H, Zhang D, et al. LncRNA KCNQ1OT1 regulates proliferation and cisplatin resistance in tongue cancer via miR-211-5p mediated Ezrin/Fak/Src signaling. Cell Death Dis. 2018;9(7):742. doi:10.1038/s41419-018-0793-5
24. Cheung M, Abu-Elmagd M, Clevers H, et al. Roles of Sox4 in central nervous system development. Mol Brain Res. 2000;79(1–2):180–191. doi:10.1016/S0169-328X(00)00109-1
25. Zhang J, Liang Q, Lei Y, et al. SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res. 2012;72(17):4597–4608. doi:10.1158/0008-5472.CAN-12-1045
26. Ruan H, Yang H, Wei H, et al. Overexpression of SOX4 promotes cell migration and invasion of renal cell carcinoma by inducing epithelial-mesenchymal transition. Int J Oncol. 2017;51(1):336–346. doi:10.3892/ijo.2017.4010
27. Li Y, Zu L, Wang Y, et al. miR-132 inhibits lung cancer cell migration and invasion by targeting SOX4. J Thorac Dis. 2015;7(9):1563.
28. Ding L, Zhao Y, Dang S, et al. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol Cancer. 2019;18(1):45. doi:10.1186/s12943-019-1006-2
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.